RNA secondary structure prediction Introduction to Part 3 of Exercise - PowerPoint PPT Presentation
1 / 7
Title:
RNA secondary structure prediction Introduction to Part 3 of Exercise
Description:
Introduction to Part 3 of Exercise ... Compute the minimum-free energy (MFE) structure ... For a given sequence: large ensemble of energetically similar structures ... – PowerPoint PPT presentation
Number of Views:530
Avg rating:3.0/5.0
Title: RNA secondary structure prediction Introduction to Part 3 of Exercise
1
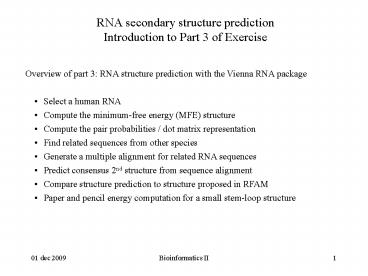
RNA secondary structure predictionIntroduction
to Part 3 of Exercise
- Overview of part 3 RNA structure prediction with
the Vienna RNA package - Select a human RNA
- Compute the minimum-free energy (MFE) structure
- Compute the pair probabilities / dot matrix
representation - Find related sequences from other species
- Generate a multiple alignment for related RNA
sequences - Predict consensus 2nd structure from sequence
alignment - Compare structure prediction to structure
proposed in RFAM - Paper and pencil energy computation for a small
stem-loop structure
2
Partition function and Equilibrium base-pair
probabilities
Motivation For a given sequence large ensemble
of energetically similar structures Base-pairs
which occur in most structures are most
interesting Definition of Partition
function F(S) is the free energy of a
partilucar secondary structure S, k is the
Boltzman constant Probability of an individual
structure Probability of a base-pair h,l
3
Dot matrix representation of Pair-probabilities
in RNA secondary structure ensemble
Upper right Pair-probalities Lower-left Pairs
in minimum free energy structure (from McCaskill
1990, Biopolymers 29, 1105.)
4
Basics of Vienna RNAalifold algorithm
- Cost function
- For loops average energy of sequences in
alignment - Base-pairs linear combination of
- average stacking energies from alignment
- modified covariance terms re-scaled as energies
(special treatment of GU pair-related
co-variance) - Constraints exclusion of pairs with many
non-standard base pairs - Folding algorithm
- Same as RNAfold since cost function is equivalent
5
Energy parameters for RNA folding
Stacking energies are determined from melting
curves of synthetic RNA hybrids. (see Walter et
al. 1994, Proc Natl Acad Sci U S A. 91, 92189222)
6
RNA secondary structure elements Terminology
7
Extended parameter system of Vienna package
Free energies to compute MFE structure at 37º
Celsius. Enthalpies serve to rescale energy
parameters for other temperatures Extended
parameters used by the Vienna package
mismatch_interiorFree energies for the
interaction between the closing pair of an
interior loop and the two unpaired bases adjacent
to the helix. mismatch_hairpinSame as above
for hairpin loops. int11_energies, (also
int21_, int22_)Sequence-dependent free energies
for interior loops dangle5Energies for the
interaction of an unpaired base on the 5' side
and adjacent to a helix dangle3Same as above
for bases on the 3' side of a helix.