Eukaryotic Gene Finding PowerPoint PPT Presentation
1 / 34
Title: Eukaryotic Gene Finding
1
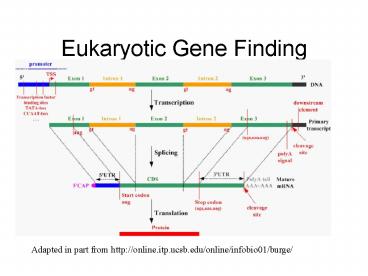
Eukaryotic Gene Finding
Adapted in part from http//online.itp.ucsb.edu/on
line/infobio01/burge/
2
Prokaryotic vs. Eukaryotic Genes
- Prokaryotes
- small genomes
- high gene density
- no introns (or splicing)
- no RNA processing
- similar promoters
- overlapping genes
- Eukaryotes
- large genomes
- low gene density
- introns (splicing)
- RNA processing
- heterogeneous promoters
- polyadenylation
3
(No Transcript)
4
Pre-mRNA Splicing
exon definition
intron definition
...
(assembly of
spliceosome,
catalysis)
...
5
(No Transcript)
6
Some Statistics
- On average, a vertebrate gene is about 30KB long
- Coding region takes about 1KB
- Exon sizes can vary from double digit numbers to
kilobases - An average 5 UTR is about 750 bp
- An average 3UTR is about 450 bp but both can be
much longer.
7
Human Splice Signal Motifs
5' splice signal
3' splice signal
8
(No Transcript)
9
(No Transcript)
10
Semi-Markov HMM Model
11
GHMM
- A finite Set Q of states
- Initial state distribution ?
- Transition probabilities Ti,j for
- Length distribution f of the states (fq is the
length distribution of state q) - Probability model for each state
12
GHMM contin.
- A parse ? of a sequence S of length L is an
ordered sequence of states (q1, . . . , qt) with
an associated duration di to each state
The most probable pass ?opt can be computed as in
Veterbi algorithm
13
Genscan HSMM
14
GenScan States
- N - intergenic region
- P - promoter
- F - 5 untranslated region
- Esngl single exon (intronless) (translation
start -gt stop codon) - Einit initial exon (translation start -gt donor
splice site) - Ek phase k internal exon (acceptor splice site
-gt donor splice site) - Eterm terminal exon (acceptor splice site -gt
stop codon) - Ik phase k intron 0 between codons 1
after the first base of a codon 2 after the
second base of a codon
15
GenScan features
- Model both strands at once
- Each state may output a string of symbols
(according to some probability distribution). - Explicit intron/exon length modeling
- Advanced splice site modeling
- Complete intron/exon annotation for sequence
- Able to predict multiple genes and partial/whole
genes - Parameters learned from annotated genes
- Separate parameter training for different CpG
content groups (lt 43, 43-51, 51-57,gt57 CG
content)
16
Various parameters in GENSCAN
17
(No Transcript)
18
GenScan Signal Modeling
- PSSM P(S) P1(S1)P2(S2) Pn(Sn)
- PolyA signal
- Translation initiation/termination signal
- Promoters
- WAM P(S) P1(S1) P2(S2S1)Pn(SnSn-1)
- 5 and 3 splice sites
19
GENSCAN Performance
- gt 80 correct exon predictions, and gt 90 correct
coding/non coding predictions by bp. - BUT - the ability to predict the whole gene
correctly is much lower
20
HMM-based Gene Finding
- GENSCAN (Burge 1997)
- FGENESH (Solovyev 1997)
- HMMgene (Krogh 1997)
- GENIE (Kulp 1996)
- GENMARK (Borodovsky McIninch 1993)
- VEIL (Henderson, Salzberg, Fasman 1997)
21
Using Sequence Similarity for Gene Finding
- Compare genomic sequence with expressed sequence
tags (ESTs) (e.g. by BLASTN), to identify regions
corresponding to processed mRNA - Compare genomic sequence to Protein DB (e.g. by
BLASTX), to identify probably coding regions - Spliced Alignment of genomic sequence of a
complete gene with a homologous protein sequence
(e.g. by PROCRUSTES) may enable exon/intron
reconstruction - Compare predicted peptides (e.g. by GENSCAN) with
protein DB to assign confidence to predictions
and functional annotations - Compare Genomic sequence with homologous from
close organisms/species (e.g. by BLAST, CLASTW),
to identify conserved regions which might
correspond to coding regions and DNA signals
22
GenomeScan
- Idea We can enhance our gene prediction by
using external information DNA regions with
homology to known proteins are more likely to be
coding exons.
- Combine probabilistic extrinsic information
(BLAST hits) with a probabilistic model of gene
structure/composition (GenScan)
- Focus on typical case when homologous but not
identical - proteins are available.
23
(No Transcript)
24
(No Transcript)
25
GeneWise Birney, Amitai
- Motivation Use good DB of protein world (PFAM)
to help us annotate genomic DNA - GeneWise algorithm aligns a profile HMM directly
to the DNA
26
Sample GeneWise Output
27
Developing GeneWise Model
- Start with a PFAM domain HMM
- Replace AA emissions with codon emissions
- Allow for sequencing errors (deletions/insertions)
- Add a 3-state intron model
28
GeneWise Model
29
GeneWise Intron Model
5 site
3 site
30
GeneWise Model
- Viterbi algorithm -gt best alignment of DNA to
protein domain - Alignment gives exact exon-intron boundaries
- Parameters learned from species-specific
statistics
31
GeneWise problems
- Only provides partial prediction, and only where
the homology lies - Does not find more genes
- Pseudogenes, Retrotransposons picked up
- CPU intensive
- Solution Pre-filter with BLAST
32
Other Sequence Usage
- Search translated genomic sequences for the
occurrences of the shot peptide motifs that are
characteristic of common protein families (e.g.
zinc finger, ATP/GTP binding modifs etc.) - Identify sequences which are probably NON coding
identify known classes of interspersed repeates
(e.g LINE SINE) in none coding regions. Can be
essential to remove these before simple BLAST is
done against ESTs.
33
Summary
- Genes are complex structures which are difficult
to predict with the required level of
accuracy/confidence - Different approaches to gene finding
- Ab Initio GenScan
- Ab Initio modified by BLAST homologies
GenomeScan - Homology guided GeneWise
34
Future Directions
- Find genes not for proteins (tRNA, rRNA, smRNA)
hard ! - Deal better with overlapping genes, multiple
genes in a single sequence - Alternative splicing/transcription/translation
a whole separate issue - The mechanisms governing it, the signals
- predicting the various genes
- Very important !