ASSIGNING GENE FUNCTION BY EXPERIMENTAL ANALYSIS PowerPoint PPT Presentation
1 / 23
Title: ASSIGNING GENE FUNCTION BY EXPERIMENTAL ANALYSIS
1
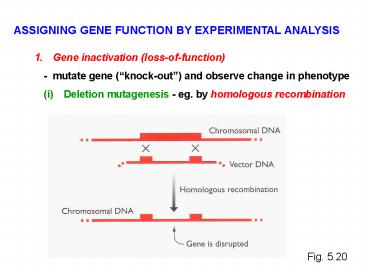
ASSIGNING GENE FUNCTION BY EXPERIMENTAL ANALYSIS
- Gene inactivation (loss-of-function)
- mutate gene (knock-out) and observe change
in phenotype
- Deletion mutagenesis - eg. by homologous
recombination
Fig. 5.20
2
Deletion cassette vector substituted DNA
can have selectable marker, restriction sites,
barcode tags
Fig. 5.30
barcode tag 20-25 nt sequence that will
uniquely identify deletion mutant is
incorporated into construct
(so can detect by hybridization or PCR)
Fig. 5.21
3
Assaying molecular barcode tags in yeast pools
- yeast deletion strains with barcodes up
downstream of KanR gene
In different environments (eg. drug D), which
strains survive? competitive fitness in
population?
Presence (abundance) of different mutant strains
monitored by bar code tags
Microarray with complementary barcode tag
sequences for all yeast genes
Steinmetz Nature Rev. Genet. 5190, 2004
So if deletion of gene X is lethal under certain
growth conditions no PCR product
4
Nature 418387, 2002
- collection of 5916 gene deletion mutants
- most showed no major phenotypic effect
Only 200 had lethal phenotype for 6 growth
conditions studied
Growth properties on galactose
Aberrant cell morphology
5
(ii) Insertional mutagenesis
Griffiths Fig. 14.18
Transposon tagging - if transposon inserts into
gene (or into regulatory sequences) gene
inactivation
Transposon tagging is random form of
mutagenesis - so prior knowledge of gene
location not required
- many different alleles can be generated
Alberts Fig. 8.55
Tn mutation in regulatory protein gene for flower
development in snapdragon
6
(iii) RNA interference
- short, antisense RNAs (21-25 nt length) in hybrid
with - specific mRNA triggers degradation
knock-down of gene expression
T7
T7
C.elegans
Alberts Fig. 8-66
Dicer ribonuclease cleaves specific mRNA into
short ds RNAs
Fig. 5.23
7
Study of 2769 C. elegans genes on chromosome 1
(p.202-203)
- in 339 cases, saw detectable change in phenotype
Emb embryonic lethal (226) Ste sterile
(96) Unc uncoordinated (70) Pep
post-embryonic
Type of gene inactivated
mature nematode
2-cell stage
660 genes required for early embryogenesis
Nature 408325, 2000
8
2. Gene over-expression (gain-of-function)
- monitor phenotypic effect of high amount of
protein
- transgenic experiments using cDNA of protein of
interest with strong promoter, high copy number
vector
Increased bone density in opg transgenic mice
Simonet Cell 89309, 1997
Fig. 5.24
9
3. Gene alteration
Site-directed mutagenesis - introduce specific
point mutation at pre-determined position
(Michael Smith UBC, Nobel prize)
5 . ATG . AAA TGT CCA . TAA 3
How to change TGT (Cys) codon to GGT (Gly) codon?
Design oligomer with mismatch to original
sequence
3 TTT CCA GGT . 5
Anneal to gene (ss form) generate copies
- using M13 phage system (p.156)
- using two-step PCR (p.157)
10
Site-directed mutagenesis using PCR
- use oligomer with mismatch as PCR primer to
generate product differing from template sequence
at desired site
Fig.T5.2
11
HOW TO DETERMINE WHERE AND WHEN GENE IS EXPRESSED?
1. Transformation of regulatory sequences
reporter gene
Fig. 5.26
Use construct with regulatory sequences for gene
of interest upstream of reporter gene such as
lacZ
b galactosidase (blue colour)
green fluorescent protein (jellyfish)
GFP
12
- can mutate regulatory sequences and monitor
phenotypic effect
Transgenic mouse embryo
Griffiths Fig. 14.27
- regulatory sequences for gene expressed in
muscle precursor cells fused to lacZ reporter
gene
13
2. Immunocytochemistry - fluorescently-tagged
antibody directed against protein of interest to
determine subcellular location
Ab for mitochondrial DNA repair protein
Mol Biol Cell 16997, 2005
Fig. 5.27
14
HOW TO STUDY PATTERNS OF GENE EXPRESSION ON LARGE
SCALE?
- to determine which sets of genes are
transcribed in certain cell type developmental
stage environmental condition drug treatment
1. RT-PCR differential display
2. SAGE serial analysis of gene expression
(Fig. 6.1)
3. DNA microarrays
4. Deep sequencing RNA (cDNA) analysis
15
1. RT-PCR differential display
Mouse heart at Embryonic days 10-16
A B 2 different sources of mRNA
Strachan Read Fig.20.8
16
2. SAGE serial analysis of gene expression
Ligate many fragments together rapid sequencing
of these concatemers
Fig. 6.1
17
TRANSCRIPT PROFILING WITH DNA MICROARRAYS
DNA chip with genes of interest (eg. clones, PCR
products, oligomer barcode tags )
- RNAs extracted from control
- and test cells (transcriptomes 1 2)
2. cDNA synthesis labeling
5cap
AAAAAAAAAn
3 5
eg. for primer can use mixture of anchored
oligo(dT)s with A, C or G in the 3 position
3. Hybridize to microarray
4. Visualize hybrids
eg. laser scanning of fluorescence
Fig. 6.3
18
Potential pitfalls with microarrays (see
p.170-171)
- if target DNA is saturated with probe,
hybridization signal strength will not reflect
mRNA abundance
Fig.6.4
- if comparing 2 transcriptomes using 2
microarrays , data must be normalized to ensure
equivalent amounts of DNA on array, same
efficiency of probe labelling, same effectiveness
of hybridization conditions....
So better to use 2 types of fluorescent probes on
one microarray
19
More efficient if transcriptomes 1 2 are
labeled with different fluorescent tags (eg
red Cy3-dUTP green Cy5-dUTP)
- then mix cDNAs and hybridize to microarray
- laser scanning ratio of fluorescence
calculated
red expressed at higher levels in test than in
control
green expressed at lower levels in test
yellow expressed at same level in both
Gibson Muse Fig. 3.1
20
TRANSCRIPT PROFILING WITH DNA MICROARRAYS
drug
No drug present
mRNAs for genes 1-3
AAAAn
AAAAn
AAAAn
AAAAn
AAAAn
AAAAn
AAAAn
RT
Red tag
Green tag
genes 1-3 on chip
21
- then cluster analysis to identify sets of
co-regulated genes
- genes with related functions tend to have
similar expression patterns
guilt-by-association
Transcriptome analysis during plant cell cycle
PNAS 9914825, 2002
- examined 1340 cell-cycle modulated genes in
tobacco
22
Some genes can give rise to more than one
distinctive mRNA
Alternative splicing
Topic 6, slide 14
mRNAs
SpliceArrays (microarray)
- using junction-specific oligomers
- different proteins can be generated from same
gene
- violates one gene one protein principle
so can have larger proteome (set of proteins)
than predicted from number of genes in genome
- may be differentially expressed
- genome-wide analyses indicate that 40-60 of
human genes have alternative splice forms
Modrek Lee 2002
Fig.6.5
23
Some applications of DNA microarrays
1. Transcript profiling (expression analysis)
2. Genotyping (SNPs)
3. Drug discovery (eg identify potential drug
targets by analyzing expression profile in
response to drug)