Comparison of Comparative Genomic Hybridization Technologies Across Microarray Platforms PowerPoint PPT Presentation
1 / 1
Title: Comparison of Comparative Genomic Hybridization Technologies Across Microarray Platforms
1
Comparison of Comparative Genomic Hybridization
Technologies Across Microarray Platforms Susan
Hester1, Laura Reid2, Agnes Viale3, Norma Nowak4,
Herbert Auer5, Kevin Knudtson6, William Ward1,
Jay Tiesman7, Caprice Rosato8, Aldo Massimi9,
Greg Khitrov10 Nancy Denslow11 1Environmental
Protection Agency, Research Triangle Park, NC
2Expression Analysis, Inc., Durham, NC 3Memorial
Sloan Kettering Cancer Center, New York, NY
4Department of Cancer Genetics, Roswell Park
Cancer Institute, Buffalo, NY 5Columbus
Childrens Hospital, Columbus, OH 6University of
Iowa, Iowa City, IA 7Procter Gamble,
Cincinnati, OH 8Center for Genome Research
Biocomputing, Oregon State University, Corvallis,
OR 9Albert Einstein College of Medicine, New
York, NY 10Department of Medicine, Mount Sinai
School of Medicine, New York, NY 11University of
Florida, Gainesville, FL
2. Accuracy HL-60 is reported to have the
following copy number changes Amplification at
8q24,Trisomy 18, monosomy X as well as Deletions
at 5q11.2-q31, 6q12, 9p21.3-p22, 10p12-p15,
14q22-q31 and17p12-p13.3. Cancer Genet Cytogenet.
2003 Nov147(1)28-35 Peiffer et al.(2006)
3. Platform Resolution Circular Binary
Segmentation (CBS) Analysis
Abstract
- Study Questions
- What is the precision of each platform?
- How accurately does each platform detect Known
Copy Number changes? - What is the resolution of microarray CGH?
Comparative Genomic Hybridization (CGH) measures
DNA copy number differences between a reference
genome and a test genome. The DNA samples are
differentially labeled and hybridized to an
immobilized substrate. In early CGH experiments,
the DNA targets were hybridized to metaphase
chromosome spreads in FISH assays. This
technology later evolved so that the DNA targets
are hybridized to microarrays containing cDNA
fragments or bacterial artificial chromosomes
(BACs). Recent commercial offerings from
Agilent, Affymetrix and Illumina derive copy
number differences using oligonucleotide
microarrays representing 500,000 or more loci. In
most commercial assays, genomic DNA is labeled
and hybridized to microarrays designed for SNP
genotyping analyses. Interestingly, Auer et al.
has recently shown that expression microarrays,
such as the Affymetrix U133 series can also be
used to identify copy number differences. In
the 2007 Association of Biomolecular Resource
Facilities (ABRF) Microarray Research Group
(MARG) project, we analyzed HL60 DNA with 5
platforms Agilent, Affymetrix 500K, Affymetrix
U133, Illumina, and RPCI BAC arrays. Data
obtained from these platforms were assessed for
precision and the ability to detect known and
novel copy number changes in the HL60 genome.
This abstract does not necessarily reflect the
views of the EPA.
Platform Chromosome Start Position End Position Number of Probes Segment Mean
RPCI BAC 5 53,817,423 139,426,041 491 -0.5315
Agilent 5 53,787,209 139,475,226 1,003 -0.1914
Illumina 5 53,729,928 113,350,609 10,438 -0.6753
AffymetrixU133 5 53,536,915 139,532,618 1212 -0.5862
Affymetrix 500K 5 53,925,665 73,549,145 13,886 -0.3785
RPCI BAC 8 98,584,025 130,781,748 14 1.2942
Agilent 8 126,357,705 130,632,571 26 0.3993
Illumina 8 126,302,381 130,771,462 428 1.1630
Affymetrix U133 8 126,335,997 128,498,243 22 1.612
Affymetrix 500K 8 126,000,000 131,000,000 1,192 1.7650
Detection of Known HL60 Gains and Losses
Platform -5q11 -6q12 8q24 -9p21 -10p12 -14q22 -17p12 Tri 18 MonoX
BAC Y N Y Y Y Y Y Y Y
Agilent Y N Y Y Y Y Y Y Y
Affy U133 Y N Y Y Y Y Y Y Y
Affy 500K Y Y Y Y Y Y Y Y Y
Illumina Y Y Y Y Y Y Y Y Y
1. Platform Precision Basis for Coefficient
of Variance Analysis- Summary statistics and
boxplot generated in Sigmaplot. BAC CV
calculated from HL60/lung ratios in 4 replicates.
Agilent CV calculated from HL60/lung ratios in
4 replicates. Illumina CV calculated from 4 R
values. Affymetrix U133 CV calculated from the
16 possible ratios between the 4 HL60 and 4 lung
replicates. Affymetrix 500K CV calculated
separately for the 250K Nsp and 250Sty GeneChips.
CV calculated from Intensities calculated from
Affymetrix software for 4 NSP CEL files and 4 STY
CEL files. These files were augmented from
Affymetrix data repository in order to calculate
expression summaries and CVs.
Detection of Novel HL60 Gains and Losses
Platform Chr.2 Chr.16 Chr.19
BAC Y Y N
Agilent N Y N
Affy U133 N Y Y
Affy 500K Y Y Y
Illumina N Y N
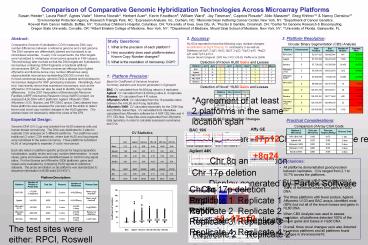
Agreement of at least 2 platforms in the same
location span
Visualizations Two Known HL60 Copy Number
Changes
Experimental Design
Practical Considerations Comparison of Array
CGH Costs
BAC 19K Display prepared from CBS Analysis of
one replicate
Genomic DNA (0.5-3 mg) was isolated from HL60
leukemia cells and human female normal lung. The
DNA was distributed to 3 sites for replicate CGH
analyses on 5 different platforms. Two platforms
used traditional (2-color) CGH methods, where
both HL60 and lung targets were hybridized to the
same microarray. Three platforms hybridized HL60
or lung targets to separate (1-color)
microarrays. Each site relied on
platform-specific protocols for target
preparation, hybridization, quality review and
copy number determination. In most cases, gains
and losses were identified based on HL60 to lung
signal ratios. For the Illumina and Affymetrix
500K platforms, gains and losses were evaluated
by comparing HL60 signals to reference datasets.
The probe annotation on all platforms was
standardized to sequence information in NCBI
build 35 (HG17).
Affy GE
Platform Microarray Price per Microarray Price per Target Labeling Process Time
RPCI BAC19K 150 175 1 day
Agilent 44K 225 180 2 days
Affymetrix 500K 250 125 2 days
Affymetrix U133 400 125 2 days
Illumina 550K 400 150 2 days
CV Statistics
-17p12
BAC Agilent Illumina Affy GE Affy Nsp Affy Sty
Mean 2.13 3.16 5.98 9.64 10.69 9.44
Median 1.96 2.66 5.23 8.71 9.91 8.47
Std Dev 1.09 2.43 3.52 4.78 5.24 5.60
Std Err 0.01 0.01 0.00 0.02 0.01 0.01
Size 18067 42897 555353 54676 524629 476709
Min 0.01 0.05 0.05 0.71 0.15 0.01
Max 16.87 77.89 98.05 59.57 48.97 50.59
Agilent 44K
8q24
Chr 8q amplification
- Conclusions
- All platforms demonstrated good precision between
replicates. CVs ranged from 2.1 to 10.7 across
the platforms. - The two platforms with the largest number of
probes, Illumina and Affymetrix 500K, identified
100 of the known losses and gains in HL60 DNA. - The three platforms with fewer probes, Agilent,
Affymetrix U133 and BAC arrays, identified most
(89) but not all of the known losses and gains
in HL60 DNA. - When CBS Analysis was used to assess resolution,
all platforms detected 100 of the known gains
and losses in HL60 DNA. - Overall, three novel changes were also detected
by various platforms and all platforms found
changes in chromosome16.
Chr 17p deletion
Platform Descriptions
Affy 500K Display generated by Partek Software
Platform Number of Probes or Probe Sets Test Site Protocol Number of Replicates Primary Data
BAC19K 19,000 in duplicate RPCI 2-color 4 HL60/lung Mean Log2 (test/lref)
Agilent 44,000 MSK 2-color 4 HL60/lung Mean Log2 (test/ref)
AffymetrixU133 55,000 CPH 1-color 4 HL60 4 lung Log2(RMA)
Affymetrix 500K 500,000 MSK 1-color 4 HL60 0 lung Reference database
Illumina 550,000 MSK 1-color 4 HL60 0 lung Log R ratio
Illumina 550K
8q24
-17p12
Replicate 1 Replicate 2 Replicate 3 Replicate 4
Replicate 1 Replicate 2 Replicate 3 Replicate 4
The test sites were either RPCI, Roswell Park
Cancer Institute MSK, Memorial Sloan-Kettering
Institute Genomic Core or CPH, Columbus
Pediatric Hospital.