MR Microimaging at 7 T: Pulse Sequence Development - PowerPoint PPT Presentation
1 / 1
Title:
MR Microimaging at 7 T: Pulse Sequence Development
Description:
In NMR spectroscopy, a sample is selected to be analyzed. For example, hydrogen nuclei have one spinning proton. Initially, these nuclei spin in different directions. – PowerPoint PPT presentation
Number of Views:53
Avg rating:3.0/5.0
Title: MR Microimaging at 7 T: Pulse Sequence Development
1
MR Microimaging at 7 T Pulse Sequence
Development Implementation
Ms. Tracy L. Doyle Mr. Ramchand
Maharaj Upper St. Clair High School, Pittsburgh,
PA Blanche Ely High School, Pompano
Beach, FL
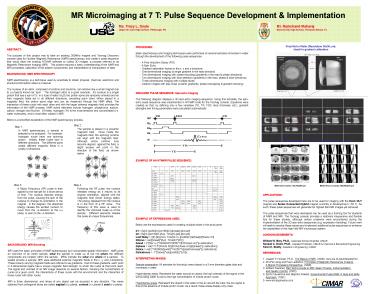
Vinyl Nut in Water (Resolution 50x50 mm)Used
for gradient calibration
- PROCEDURE
- Basic spectroscopy and imaging techniques were
performed on several samples immersed in water
through the development of the following pulse
sequences. - - A Free Induction Decay (FID)
- - A Spin Echo
- - Gradient calibration factors in the x, y and z
directions - - One-dimensional imaging (a single gradient in
the read direction) - Two-dimensional imaging with phase encoding
(gradients in the read phase directions) - - Two-dimensional imaging with slice selection
(gradients in the read, phase slice directions) - Three-dimensional imaging with multiple slices
- Cleaner images with less noise (crusher
gradients, phase rewrapping gradient ramping)
- IDEALIZED PULSE SEQUENCE Spin-echo Imaging
- The following diagram displays a 3D spin-echo
imaging sequence. Using this template, the
spin-echo pulse sequence was implemented in NTNMR
code for the Tecmag console. Equations were
created so that, by defining only a few variables
(TE, TR, FOV, slice thickness, etc.), gradient
strengths and timing parameters were calculated
automatically.
AP
PLICATIONS The pulse sequences developed here
are to be used for imaging with the Keck 25-T
magnet and Series Connected Hybrid magnet
currently in development (36 T). As such, these
pulse sequences will generate the highest field
MR images yet achieved. The pulse sequences that
were developed can be used as a training tool for
students of NMR and MRI. The Tecmag console
provides a relatively inexpensive and flexible
tool for these studies, although certain problems
were encountered during the implementation of the
3D spin-echo sequences (e.g. expression
handling). Future work will seek to resolve these
issues and implement additional pulse sequences
to enhance the capabilities of this high field MR
microscopy system. ACKNOWLEDGEMENTS William W.
Brey, Ph.D., Associate Scholar Scientist,
CIMAR Samuel C. Grant, Ph.D., Assistant
Professor, CIMAR Chemical Biomedical
Engineering Kiran K. Shetty, Assistant in
Engineering, CIMAR REFERENCES 1. Joseph P.
Hornak, Ph.D., The Basics of MRI, (2006).
www.cis.rit.edu/htbooks/mri/ 2. Zhi-Pei Liang and
Paul Lauterbur, Principles of Magnetic Resonance
Imaging A Signal Processing Perspective,
(1996). 3. William Faulkner, Rad Techs Guide to
MRI Basic Physics, Instrumentation, and
Quality Control, (2002). 4. Eiichi Fukushima and
Stephen Roeder, Experimental Pulse NMR A Nuts
and Bolts Approach, (1981). 5.
www.simplyphysics.com
ABSTRACT The purpose of this project was to
take an existing 300MHz magnet and Tecmag
Discovery console used for Nuclear Magnetic
Resonance (NMR) spectroscopy, and create a pulse
sequence that would allow the existing NTNMR
software to collect 3D images, a process referred
to as Magnetic Resonance Imaging (MRI). This
project required a basic understanding of the NMR
and MRI processes, calibration of the system
components, and interpretation manipulation of
data. BACKGROUND NMR SPECTROSCOPY NMR
spectroscopy is a technique used by scientists to
obtain physical, chemical, electronic and
structural information about a molecule. The
nucleus of an atom, composed of protons and
neutrons, can behave like a small magnet due to a
property known as spin. The hydrogen atom is a
good example. Its nucleus is a single proton
that has a spin of ½. In a tube of water (H2O)
the proton spins are randomly oriented and so
their magnetic fields act in all different
directions, cancelling each other. When placed in
a magnetic field, the proton spins align and can
be measured through the NMR effect. The
interaction of these nuclei with each other and
with the larger external magnetic field provides
the information of the NMR process. NMR active
elements include hydrogen, phosphorus, sodium,
carbon, nitrogen and fluorine. Of these,
hydrogen (1H) is the most sensitive and
concentrated (i.e. water molecules), and is most
often utilized in MRI. Below is a simplified
explanation of the NMR spectroscopy process.
1 mm
1 mm
Multiple Sagittal Slices of Vinyl Screw
(50x50x370 mm)
Step 2 The sample is placed in a powerful
magnetic field. Once inside the magnetic field,
the spinning protons will align with the magnetic
field. Although some protons may become aligned
against the field, a slight excess will point in
the direction of the field, as shown below.
Step 1 In NMR spectroscopy, a sample is
selected to be analyzed. For example, hydrogen
nuclei have one spinning proton. Initially,
these nuclei spin in different directions. The
different spins create different magnetic fields
in a variety of directions.
Zebra Fish Imaged at 11.75 T using Bruker
software console
Step 3 A Radio Frequency (RF) pulse is then
applied to the sample for a short period of time.
The nucleus absorbs energy from the pulse,
causing the spin of the nucleus to change its
orientation in the magnet. In the diagram, the
absorbed energy causes the excited nucleon to
move from the z-direction to the x-y plane, or
even to the z-direction.
Step 4 Following the RF pulse, the nucleus
releases energy as it returns to its original
orientation, aligned with the magnetic field
(lower energy state). The energy released from
the nucleus is in the form of a RF wave. The
frequency of this wave is used to identify the
chemical make up of the sample. Different
elements release this pulse at unique frequencies.
Multi-slice axials 78x78x500 mm
Multi-slice coronals 86x86x250 mm
BACKGROUND MRI Encoding MRI uses the basic
principles of NMR spectroscopy but incorporates
spatial information. NMR gives an overview of an
entire sample, what the sample is made up of but
not where the different components are located
within the sample. MRIs indicate the what and
where of a sample. To spatial encode a sample,
MRI uses additional external magnetic fields in
the x, y, and z-directions. These linearly
varying magnetic fields are referred to as
gradients. Due to these gradients, each point in
3-dimensional space has a unique magnetic field
strength, to which the nuclei at that point
react. The signal and contrast of an MR image
depends on several factors, including the
concentration of nuclei at a given point, the
interactions of those nuclei with the environment
and the interaction of those nuclei with other
nuclei. MRI is three dimensional, and slices of
any object can be acquired in any direction. The
views obtained from orthogonal slices are called
sagittal (z-y plane), coronal (z-x plane) axial
(x-y plane).