Regulating%20gene%20expression - PowerPoint PPT Presentation
Title:
Regulating%20gene%20expression
Description:
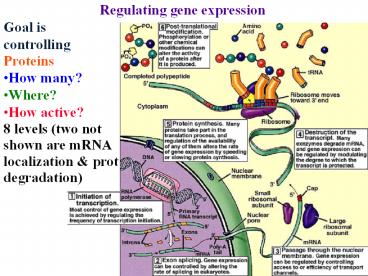
Regulating gene expression Goal is controlling Proteins How many? Where? How active? 8 levels (two not shown are mRNA localization & prot degradation) – PowerPoint PPT presentation
Number of Views:281
Avg rating:3.0/5.0
Title: Regulating%20gene%20expression
1
- Regulating gene expression
- Goal is
- controlling
- Proteins
- How many?
- Where?
- How active?
- 8 levels (two not
- shown are mRNA
- localization prot
- degradation)
2
- Transcription in Eukaryotes
- Pol I only makes 45S-rRNA precursor
- 50 of total RNA synthesis
- insensitive to ?-aminitin
- Mg2 cofactor
- Regulated _at_ initiation frequency
3
RNA Polymerase III makes ribosomal 5S and tRNA
( some snRNA scRNA) gt100 different kinds of
genes 10 of all RNA synthesis Cofactor Mn2
cf Mg2 sensitive to high ?-aminitin
4
- RNA Polymerase II
- makes mRNA (actually hnRNA), some snRNA and scRNA
- 30,000 different gene models
- 20-40 of all RNA synthesis
- very sensitive to ?-aminitin
5
Initiation of transcription by Pol II Basal
transcription 1) TFIID binds TATAA box 2) TFIIA
and TFIIB bind to TFIID/DNA 3) Complex recruits
Pol II 4) Still must recruit TFIIE TFIIH to
form initiation complex
6
Initiation of transcription by Pol II Basal
transcription 1) Once assemble initiation complex
must start Pol II 2) Kinase CTD negative charge
gets it started 3) Exchange initiation for
elongation factors 4) Continues until hits
terminator
7
- Initiation of transcription by Pol II
- Basal transcription
- 1) Once assemble initiation complex must start
Pol II - 2) Kinase CTD
- negative charge
- gets it started
- 3) RNA pol II is paused
- on many promoters!
- even of genes that
- arent expressed!
- Early elongation is also
- regulated!
8
- Initiation of transcription by Pol II
- RNA pol II is paused on many promoters!
- even of genes that arent expressed! (low
mRNA) - Early elongation is also
- regulated!
- PTEFb kinases CTD to
- stimulate processivity
- processing
9
- Initiation of transcription by Pol II
- RNA pol II is paused on many promoters!
- even of genes that arent expressed! (low
mRNA) - Early elongation is also
- regulated!
- PTEFb kinases CTD to
- stimulate processivity
- processing
- Many genes have
- short transcripts
10
- Initiation of transcription by Pol II
- RNA pol II is paused on many promoters!
- even of genes that arent expressed! (low
mRNA) - Early elongation is also
- regulated!
- PTEFb kinases CTD to
- stimulate processivity
- processing
- Many genes have
- short transcripts
- Yet another new
- level of control!
11
Transcription Template strand determines next
base Positioned by H-bonds until RNA
polymerase links 5 P to 3 OH in front
12
Transcription Template strand determines next
base Positioned by H-bonds until RNA
polymerase links 5 P to 3 OH in front Energy
comes from hydrolysis of 2 Pi
13
Transcription NTP enters E site rotates into A
site
14
Transcription NTP enters E site rotates into A
site Specificity comes from trigger loop
15
Transcription Specificity comes from trigger
loop Mobile motif that swings into position
triggers catalysis
16
Transcription Specificity comes from trigger
loop Mobile motif that swings into position
triggers catalysis Release of PPi triggers
translocation
17
Transcription Proofreading when it makes a
mistake it removes 5 bases tries again
18
Activated transcription by Pol II Studied by
mutating promoters for reporter genes
19
Activated transcription by Pol II Studied by
mutating promoters for reporter genes Requires
transcription factors and changes in chromatin
20
- Activated transcription by Pol II
- enhancers are sequences 5 to TATAA
- transcriptional activators bind them
- have distinct DNA binding and activation domains
21
- Activated transcription by Pol II
- enhancers are sequences 5 to TATAA
- transcriptional activators bind them
- have distinct DNA binding and activation domains
- activation domain interacts with mediator
- helps assemble initiation complex on TATAA
22
- Activated transcription by Pol II
- enhancers are sequences 5 to TATAA
- transcriptional activators bind them
- have distinct DNA binding and activation domains
- activation domain interacts with mediator
- helps assemble initiation complex on TATAA
- Recently identified activating RNA bind
enhancers mediator
23
- Activated transcription by Pol II
- Other lncRNA promote transcriptional poising in
yeast - http//www.plosbiology.org/article/info3Adoi2F10
.13712Fjournal.pbio.1001715 - lncRNA displaces
- glucose-responsive
- repressors co-
- repressors from genes
- for galactose catabolism
24
- Activated transcription by Pol II
- Other lncRNA promote transcriptional poising in
yeast - http//www.plosbiology.org/article/info3Adoi2F10
.13712Fjournal.pbio.1001715 - lncRNA displaces
- glucose-responsive
- repressors co-
- repressors from genes
- for galactose catabolism
- Speeds induction of
- GAL genes
25
Euk gene regulation Initiating transcription is
1st most important control Most genes are
condensed only express needed genes not enough
room in nucleus to access all genes at same
time! must find decompress gene
26
- First remodel chromatin
- some proteins reposition
- nucleosomes
- others acetylate histones
- Neutralizes ve charge
- makes them release DNA
27
- Epigenetics
- heritable chromatin modifications are associated
with activated repressed genes
28
Epigenetics ChIP-chip ChiP-seq data for whole
genomes yield complex picture 17 mods are
associated with active genes in CD-4 T cells
29
- Generating methylated DNA
- Si RNA are key generated from antisense or
foldbackRNA
30
- Generating methylated DNA
- Si RNA are from antisense or foldback RNA
- Primary 24 nt siRNA are generated by DCL3
31
- Generating methylated DNA
- Si RNA are from antisense or foldback RNA
- Primary 24 nt siRNA are generated by DCL3
somehow polIV is attracted to make more RNA
32
- Generating methylated DNA
- Si RNA are from antisense or foldback RNA
- Primary 24 nt siRNA are generated by DCL3
somehow polIV is attracted to make more RNA - RDR2 makes bottom strand
33
- Generating methylated DNA
- Si RNA are from antisense or foldback RNA
- Primary 24 nt siRNA are generated by DCL3
somehow polIV is attracted to make more RNA - RDR2 makes bottom strand
- DCL3 cuts dsRNA into 24nt
- 2 siRNA
34
- Generating methylated DNA
- Si RNA are from antisense or foldback RNA
- Primary 24 nt siRNA are generated by DCL3
somehow polIV is attracted to make more RNA - RDR2 makes bottom strand
- DCL3 cuts dsRNA into 24nt
- 2 siRNA
- Amplifies signal!-gt extends
- Methylated region
35
- Generating methylated DNA
- Si RNA are from antisense or foldback RNA
- Primary 24 nt siRNA are generated by DCL3
somehow polIV is attracted to make more RNA - RDR2 makes bottom strand
- DCL3 cuts dsRNA into 24nt
- 2 siRNA
- Amplifies signal!-gt extends
- Methylated region
- These guide silencing
- Complex to target site
- (includes Cytosine H3K9
- Methyltransferases)
36
mRNA PROCESSING Primary transcript is
hnRNA undergoes 3 processing reactions before
export to cytosol All three are coordinated with
transcription affect gene expression enzymes
piggy-back on POLII
37
mRNA PROCESSING Primary transcript is
hnRNA undergoes 3 processing reactions before
export to cytosol 1) Capping addition of 7-methyl
G to 5 end
38
mRNA PROCESSING Primary transcript is
hnRNA undergoes 3 processing reactions before
export to cytosol 1) Capping addition of 7-methyl
G to 5 end identifies it as mRNA needed for
export translation
39
mRNA PROCESSING Primary transcript is
hnRNA undergoes 3 processing reactions before
export to cytosol 1) Capping addition of 7-methyl
G to 5 end identifies it as mRNA needed for
export translation Catalyzed by CEC attached to
POLII
40
- mRNA PROCESSING
- 1) Capping
- 2) Splicing removal of introns
- Evidence
- electron microscopy
- sequence alignment
41
Splicing the spliceosome cycle 1) U1 snRNP
(RNA/protein complex) binds 5 splice site
42
SplicingThe spliceosome cycle 1) U1 snRNP binds
5 splice site 2) U2 snRNP binds
branchpoint -gt displaces A at branchpoint
43
SplicingThe spliceosome cycle 1) U1 snRNP binds
5 splice site 2) U2 snRNP binds
branchpoint -gt displaces A at branchpoint 3)
U4/U5/U6 complex binds intron displace
U1 spliceosome has now assembled
44
Splicing RNA is cut at 5 splice site cut end
is trans-esterified to branchpoint A
45
Splicing 5) RNA is cut at 3 splice site 6) 5
end of exon 2 is ligated to 3 end of exon 1 7)
everything disassembles -gt lariat intron is
degraded
46
SplicingThe spliceosome cycle
47
Splicing Some RNAs can self-splice! role of
snRNPs is to increase rate! Why splice?
48
Splicing Why splice? 1) Generate diversity
exons often encode protein domains
49
Splicing Why splice? 1) Generate
diversity exons often encode protein
domains Introns larger target for insertions,
recombination
50
Why splice? 1) Generate diversity gt94 of human
genes show alternate splicing
51
Why splice? 1) Generate diversity gt94 of human
genes show alternate splicing same gene encodes
different protein in different tissues
52
Why splice? 1) Generate diversity gt94 of human
genes show alternate splicing same gene encodes
different protein in different tissues Stressed
plants use AS to make variant stress-response
proteins
53
Why splice? 1) Generate diversity gt94 of human
genes show alternate splicing same gene encodes
different protein in different tissues Stressed
plants use AS to make variant Stress-response
proteins Splice-regulator proteins control AS
regulated by cell-specific expression and
phosphorylation
54
Splicing Why splice? 1) Generate diversity 2)
Modulate gene expression introns affect amount of
mRNA produced
55
mRNA Processing RNA editing Two types C-gtU and
A-gtI
56
- mRNA Processing RNA editing
- Two types C-gtU and A-gtI
- Plant mito and cp use C -gt U
- gt300 different editing events have been detected
in plant mitochondria some create start stop
codons
57
- mRNA Processing RNA editing
- Two types C-gtU and A-gtI
- Plant mito and cp use C -gt U
- gt300 different editing events have been detected
in plant mitochondria some create start stop
codons way to prevent nucleus from stealing
genes!
58
- mRNA Processing RNA editing
- Human intestines edit APOB mRNA C -gt U to create
a stop codon _at_ aa 2153 (APOB48) cf full-length
APOB100 - APOB48 lacks the CTD LDL receptor binding site
59
- mRNA Processing RNA editing
- Human intestines edit APOB mRNA C -gt U to create
a stop codon _at_ aa 2153 (APOB48) cf full-length
APOB100 - APOB48 lacks the CTD LDL receptor binding site
- Liver makes APOB100 -gt correlates with heart
disease
60
- mRNA Processing RNA editing
- Two types C-gtU and A-gtI
- Adenosine de-aminases (ADA) are ubiquitously
expressed in mammals - act on dsRNA convert A to I (read as G)
61
- mRNA Processing RNA editing
- Two types C-gtU and A-gtI
- Adenosine de-aminases (ADA) are ubiquitously
expressed in mammals - act on dsRNA convert A to I (read as G)
- misregulation of A-to-I RNA editing has been
implicated in epilepsy, amyotrophic lateral
sclerosis depression
62
- mRNA Processing Polyadenylation
- Addition of 200- 250 As to end of mRNA
- Why bother?
- helps identify as mRNA
- required for translation
- way to measure age of mRNA
- -gtmRNA s with lt 200 As have short half-life
63
- mRNA Processing Polyadenylation
- Addition of 200- 250 As to end of mRNA
- Why bother?
- helps identify as mRNA
- required for translation
- way to measure age of mRNA
- -gtmRNA s with lt 200 As have short half-life
- gt50 of human mRNAs have alternative polyA sites!
64
mRNA Processing Polyadenylation gt50 of human
mRNAs have alternative polyA sites!
65
- mRNA Processing Polyadenylation
- gt50 of human mRNAs have alternative polyA sites!
- result different mRNA, can result in altered
export, stability or different proteins
66
- mRNA Processing Polyadenylation
- gt50 of human mRNAs have alternative polyA sites!
- result different mRNA, can result in altered
export, stability or different proteins - some thalassemias are due to mis-poly A
67
mRNA Processing Polyadenylation some
thalassemias are due to mis-poly A Influenza
shuts down nuclear genes by preventing
poly-Adenylation (viral protein binds CPSF)
68
mRNA Processing Polyadenylation 1) CPSF
(Cleavage and Polyadenylation Specificity Factor)
binds AAUAAA in hnRNA
69
mRNA Processing Polyadenylation 1) CPSF binds
AAUAAA in hnRNA 2) CStF (Cleavage Stimulatory
Factor) binds G/U rich sequence 50 bases
downstream CFI, CFII bind in between
70
Polyadenylation 1) CPSF binds AAUAAA in hnRNA 2)
CStF binds CFI, CFII bind in between 3) PAP
(PolyA polymerase) binds cleaves 10-35 b 3 to
AAUAAA
71
mRNA Processing Polyadenylation 3) PAP (PolyA
polymerase) binds cleaves 10-35 b 3 to
AAUAAA 4) PAP adds As slowly, CFI, CFII and CPSF
fall off
72
- mRNA Processing Polyadenylation
- 4) PAP adds As slowly, CFI, CFII and CPSF fall
off - PABII binds, add
- As rapidly until 250
73
Coordination of mRNA processing Splicing and
polyadenylation factors bind CTD of RNA Pol II-gt
mechanism to coordinate the three
processes Capping, Splicing and Polyadenylation
all start before transcription is done!
74
Export from Nucleus Occurs through nuclear
pores anything gt 40 kDa needs
exportin protein bound to 5 cap
75
- Export from Nucleus
- In cytoplasm nuclear proteins fall off, new
proteins bind - eIF4E/eIF-4F bind cap
- also new
- proteins bind
- polyA tail
- mRNA is
- ready to be
- translated!