MAXIMUM LIKELIHOOD ESTIMATION - PowerPoint PPT Presentation
1 / 19
Title:
MAXIMUM LIKELIHOOD ESTIMATION
Description:
1. MAXIMUM LIKELIHOOD ESTIMATION. Recall general discussion ... Also defined the Log-likelihood (Support function S( ) ) and its ... fits a parabola to L.F. ... – PowerPoint PPT presentation
Number of Views:67
Avg rating:3.0/5.0
Title: MAXIMUM LIKELIHOOD ESTIMATION
1
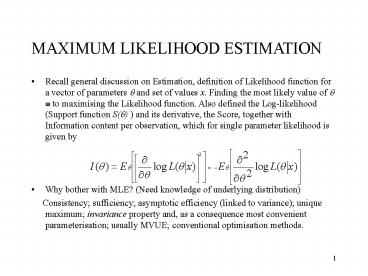
MAXIMUM LIKELIHOOD ESTIMATION
- Recall general discussion on Estimation,
definition of Likelihood function for a vector of
parameters ? and set of values x. Finding the
most likely value of ? ? to maximising the
Likelihood function. Also defined the
Log-likelihood (Support function S(?) ) and its
derivative, the Score, together with Information
content per observation, which for single
parameter likelihood is given by - Why bother with MLE? (Need knowledge of
underlying distribution) - Consistency sufficiency asymptotic
efficiency (linked to variance) unique maximum
invariance property and, as a consequence most
convenient parameterisation usually MVUE
conventional optimisation methods.
2
Estimator Comparison in brief.
- Classical, uses objective probabilities,
intuitive estimators, additional assumptions for
sampling distributions, good properties for some
estimators. (See LSE) - Moment - less calculation, loss of efficiency.
Not that widely used in genomic analysis even
though usually have analytical solutions and low
bias, because poorer asymptotic properties and
even simple solutions may not be unique. - Bayesian - subjective prior knowledge, sample
info. close to MLE under certain conditions - see
earlier. - LSE - if assumptions OK, ?s unbiased variances
obtained (XTX)-1 Assumptions needed on
distributions of response variables are just
expectations and variance-covariance structure.
(Unlike MLE where need to specify joint prob.
distribution of variables). But additional
assumptions for sampling distns. Some
computational advantage. Close if assumptions met
e.g. in Likelihood form, LSE conditions
3
VARIANCE, BIAS and CONFIDENCE INTERVALS
- Variance of an Estimator - usual form or
- for k independent estimates
- For a large sample, variance of an MLE can be
approximated by - can also be estimated empirically, using
re-sampling techniques. - Variance of a linear function of several
estimates - common in statistical genomics, see
earlier. - Recall Bias of the Estimator
- then the Mean Square error is defined to
be - expands to
- so we have the basis for C.I. and tests of
hypothesis.
4
COMMONLY-USED METHODS of obtaining MLE
- Analytical - solving or
when simple solutions exist - Grid search or likelihood profile approach
- Newton-Raphson iteration methods
- EM (expectation and maximisation) algorithm
- N.B. Log.-likelihood, because maximum for same
value of ? as - Likelihood
- Easier to compute
- Close relationship between statistical
properties of MLE and Log- - likelihood
5
METHODS in brief
- Analytical - recall Binomial example earlier
- Example For Normal, MLEs of mean and variance,
(taking derivatives w.r.t mean and variance
separately), and equivalent to sample mean and
actual variance (i.e. /N), -unbiased if mean
known, biased if not. - Invariance One-to-one relationships preserved
- Used when MLE has a simple solution
6
Methods for MLEs contd.
- Grid Search MLE from plots likelihood/
log-likelihood vs parameter. - Relative Likelihood Likelihood/Max. Likelihood
(set 1). - Peak of R.L. can be visually identified or
from searching algorithm. E.g. suppose - -Plot likelihood -parameter space range
- gives 2 peaks, - symmetrical around ?
likelihood profile for the well-known - mixed linkage phase problem in linkage
analysis. If constrain - MLE R.F. between genes
(possible mixed linkage phase). - Graphic/numerical Implementation -initial
estimate of ?, direction of search determined by
evaluating likelihood at both sides of ?. Search
takes direction of increase. Initial search
increments large, e.g. 0.1, then when likelihood
starts to decrease, stop and refine increment. - Multiple peaks - miss global maximum,
computationally intensive - Multiple Parameters - grid search. Interpretation
of Likelihood profiles can be difficult.
7
Example
- Recall Exercises 2, ex. 8. Data used to show a
linkage relationship between marker and a
rust-resistantgene. Escapes individuals who
are susceptible, but show no disease (rust)
phenotype under experimental conditions. So
define as proportion escapes and R.F.
respectively. is penetrance for disease
trait, i.e. Pindividual with susceptible
genotype has disease phenotype. Purpose of this
type of experiment typically to estimate R.F.
between marker and gene. - Support function
- Setting first derivatives w.r.t 0.
No simple analytical solution - Using grid search, likelihood reaches maximum at
- In general, this type of experiment tests H0
Independence between marker and gene
and no escapes using Likelihood Ratio
Test statistics. - N.B for Moment estimates (ex. 7) solve
- - not same as MLE
8
Methods contd.
- Newton-Raphson Iteration
- Have Score (?) 0 from previously. N-R consists
of replacing Score by linear terms of its Taylor
expansion, so if ? a solution, ? first guess -
Repeat with ? replacing ? -
Each iteration - fits a parabola to L.F. - Problems -Multiple peaks, zero Information,
extreme estimates - Multiple parameters - matrix notation, where S
matrix for example has elements derivatives of
S(?, ?) w.r.t. ? and ? respectively. Similarly,
the Information matrix has terms of form -
? Estimates -
are
9
Methods contd.
- Expectation-Maximisation Algorithm - Iterative.
Incomplete data - (Much genomic data fits this situation e.g.
linkage analysis with marker genotypes of F2
progeny, usually 9 categories observed for
2-locus, 2-allele model, but 16 complete info.,
while 14 give info. on linkage. Some hidden, but
if linkage parameter known, expected frequencies
can be predicted and the complete data restored
by expectation). - Steps - Expectation estimates statistics of
complete data, given observed incomplete data.
Maximisation uses estimated complete data to give
MLE. Iterate till converges. - Implementation An initial guess ? chosen (e.g.
0.25 say for R.F.). Taking this true,
complete data estimated, by distributional
statements e.g. P(individual is recombinant given
observed genotype) for R.F. estimation. MLE
estimate ? computed. This, for R.F. ? sum of
recombinants/N. Thus MLE, for fi observed count,
-
Convergence ? ? or
10
LIKELIHOOD for C.I. and H.T.
- Likelihood Ratio Test - cf with ?2. Principal
Advantage of G is Power, where unknown parameters
involved in hypothesis test. - Have likelihood of ?
taking a value ?A which maximises - it, i.e. MLE and likelihood ? under H0 ?N
, e.g. ?N 0.5 - Form of L.R. Test Statistic
-
or, conventionally - In practice - interpretation issue - choose
to use first form. - Distribution of G approx. ?2 - dof
difference in dimension of -
parameter spaces for L(?A), L(?N) - Goodness of Fit
.notation as for ?2 , G ?2n-1 - Independence
notation, dof as ?2
11
Example
- To test H0 ? 0.5 (estimated parameter of
Binomial) - H1 ? ? 0.5
- where is MLE of Binomial parameter. If
and x replaced with expectations or
parametric values - i.e. expected Likelihood Ratio test statistic
sample size n , parameter ? - where the part in the bracket is the ELRTS
from a single observation
12
Power-Example extended
- Under H0
- At level of significance ?0.05, suppose true ?
? 1 0.2, so if n25 - (in genomics might apply where R.F. 0.2
between two genes (as opposed to 0.5). Natural
logs. used, though either possible in practice.
Hence, generic form Log rather than Ln here.
Assume Ln throughout unless otherwise indicated) - Rejection region at 0.05 level is
- If sketch the curves, PLRTS falls in the
acceptance region 0.13, - the probability of a false negative when
actual value of ? 0.2 - If sample size increased, e.g. n50, EG 19
and easy to show that PFalse negative 0.01 - Generally Power for these tests given by
13
Likelihood Confidence Intervals -method
- Example Consider the following Likelihood
function - where ? is the unknown parameter and a, b
observed counts - If four sets of data observed,
- A (a,b) (8,2), B (a,b)(16,4) C
(a,b)(80, 20) D (a,b) (400, 100) - Likelihood estimates can be plotted vs
possible parameter values, with MLE peak value.
For example, MLE 0.2, Lmax0.0067 for A,
0.0045 for B etc. - ? A Log Lmax - Log LLog (0.0067)-Log(0.00091)
2 gives ? 95 C.I. - and ? (0.035,0.496) corresponding to
L0.00091, ? 95 C.I. for A. - Similarly, manipulating this expression,
Likelihood value corresponding to ? 95
confidence interval given as - L 7.389Lmax
- Usually plot Log-likelihood vs parameter, rather
than Likelihood - As sample size increases, interval narrower and ?
symmetric
14
Example - sample size
- For expected Log-LRTS
and average Info. content (per observation) -
-
- If true parameter values 0.05,0.1, 0.2, 0.3
respectively, then - ? G I(?) and sample
size for power 90 (1- ? 0.9) and - 0.05 0.99 21.1 ? 0.05 from?
- 0.10 0.74 11.1
- 0.20 0.39 6.3 ? so have ?
- 0.30 0.17 4.8 ? Size
? or if want, say, range (d) of CI - 0.05 11
? true value of parameter, - 0.10 15
(i.e. d ? ?) - c.f. classical form - 0.20 28
- 0.30 64
15
Multiple Populations Extensions to G -Example
- Recall Mendels data - Week 3 and Extensions to
?2 - Week 8. - In brief Round Wrinkled
- Plant O E O
E G dof p-value - 1 45 42.75 12
14.25 0.49 1 0.49 - 2
0.09 1 0.77 - 3
0.10 1 0.75 - 4
1.30 1 0.26 - 5
0.01 1 0.93 - 6
0.71 1 0.40 - 7
0.79 1 0.38 - 8
0.63 1 0.43 - 9
1.06 1 0.30 - 10
0.17 1 0.68 - Total 336 101
5.34 10 - Pooled 336 327.75 101 109.25
0.85 1 0.36 - Heterogeneity
4.50 9 0.88
16
Multiple Populations - summary
- Parallels
- Partitions therefore
- and Gheterogeneity Gtotal - GPooled
(nno. classes, p no. populations) - Example in brief Recall Backcross (AaBb x aabb)
-Goodness of fit etc. (2- locus model),Week 3.
For each of the four crosses, a Total GoF
statistic can be calculated according to expected
segregation ratio 1111 - assumes no
segregation distortion for both loci and no
linkage between loci. For each locus GoF
calculated using marginal counts, assuming the
two genotypes segregate 11.Difference between
Total and 2 individual locus GoF statistics is
L-LRTS (or chi-squared statistic) contributed by
association/linkage between 2 loci.
17
Class Exercise solutions
- Mendels Peas Week 3 - ?2 - extensions, Week 8
- In brief Round Wrinkled
- Plant O E O
E ?2 dof p-value - 1 45 42.75 12
14.25 0.47 1 0.49 - 2
0.09 1 0.77 - 3
0.10 1 0.75 - 4
1.39 1 0.24 - 5
0.01 1 0.93 - 6
0.67 1 0.41 - 7
0.76 1 0.38 - 8
0.67 1 0.41 - 9
0.98 1 0.32 - 10
0.17 1 0.68 - Total 336 101
5.30 10 - Pooled 336 327.75 101 109.25
0.83 1 0.36 - Heterogeneity
4.47 9 0.88 - No significant departure from the expected
frequencies detected for each of the 10 plants or
for the pooled frequencies. The heterogeneity ?2
also not significant. Notes - separate H0. Some
differences in ?2 , compared to G values
(Lecture)
18
Class Examples contd.
- Two-way ANOVA/Additive Design, Week 8, - solution
in lecture - Backcross (Wk 3 referred to Wk 10) - Complete
GoF etc. ?2 analysis - Cross Total Locus A Locus
B Linkage - 1 2.13 0.06 (0.86)
0.01(0.91) 2.09(0.15) p-values
in brackets - 2 6.60 0.03(0.86)
0.03(0.86) 6.53(0.01) - 3 66.00 0.33(0.56)
0.33(0.56) 65.33(lt0.0001) - 4 11.60 0.27(0.61)
0.07(0.80) 11.27(0.0008) - Total 86.33 0.66
0.45 85.22 Each
cross ?12, Total - Pooled 61.86 0.15(0.70)
0.33(0.56) 61.38(lt0.0001) Sum of 4 crosses - Heterogeneity 24.47 0.51(0.92) 0.12(0.99)
23.84(lt0.0001) - Pooled - uses marginal frequency of 4 genotypic
classes over 4 crosses (Assumes no heterogeneity
in Segregation Ratio among 4 crosses - for each
locus and for linkage relationship between them).
Locus A, B and Linkage ? ?32 under (different)Ho - Heterogeneity overall ?92 where dof from
(4-1)? (4-1) under H0 - CONCLUSIONS -No S.R. distortion for 2 loci (all
4 crosses) - - Significant linkage in 3 crosses (2,3,4)
- -Significant Heterogeneity among 4 crosses found
for linkage relationship between 2 loci. - -Sig.GoF statistic for heterogeneity mainly from
Cross 1 compared with others, thus linkage - p-value for heterogeneity GoF from 2,3,4 as
above - Experimentally ?, Cross 1 biologically different
from others, so linkage between loci A and B
could not be detected using cross 1 data
19
Outstanding class exercises
- Likelihood C.I. for data sets B,C,D - Lectures
Week 10 - Sample size calculations for range ? true
parameter values given - Lectures Week 10 - Backcross example - to complete for G to compare
with ?2 results - (Week 3, Week 8 and current)