Review: Amino Acid Side Chains - PowerPoint PPT Presentation
1 / 14
Title:
Review: Amino Acid Side Chains
Description:
Review: Amino Acid Side Chains. Aliphatic- Ala, Val, Leu, Ile, Gly ... Side chains collision also limit f/ combinations. Backbone restricted Secondary structure ... – PowerPoint PPT presentation
Number of Views:65
Avg rating:3.0/5.0
Title: Review: Amino Acid Side Chains
1
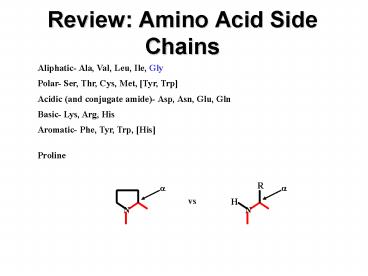
Review Amino Acid Side Chains
Aliphatic- Ala, Val, Leu, Ile, Gly Polar- Ser,
Thr, Cys, Met, Tyr, Trp Acidic (and conjugate
amide)- Asp, Asn, Glu, Gln Basic- Lys, Arg,
His Aromatic- Phe, Tyr, Trp, His Proline
2
Review Backbone Conformation
- Side chains collision also limit f/?
combinations - Backbone restricted ? Secondary structure limited
3
Review Heirarchy of Structure
Primary- sequence Secondary- local Supersecondary
(motifs)- intermediate Domains- independent
folding units Tertiary- organization of a
complete chain Quaternary- organization of
multiple chains
4
Review Tertiary Structure
- Soluble proteins have an inside (core) and
outside - Folding driven by water- hydrophilic/phobic
- Side chain properties specify core/exterior
- Some interactions inside, others outside
- Specific structures result from side chain
interactions - Hydrophobic interactions (interior)
- Hydrogen bonds (interior and exterior)
- Ionic Interactions (exterior)
5
Relationships Among Proteins
- Many sequences can give same tertiary
structure - Side chain pattern more important than sequence
- When sequence homology is high (gt50),
probably same structure and function (structural
genomics) - Cores conserved
- Surfaces and loops more variable
- 3-D shape more conserved than sequence
- There are a limited number of structural
frameworks
6
Relationships Among Proteins
- I. Homologous conserved sequence (cytochrome c)
- Same structure
- Same function
- Modeling structure from homology
- II. Similar function- different sequence
(dehydrogenases) - One domain same structure
- One domain different
- III. Similar structure- different function (cf.
thioredoxin) - Same 3-D structure
- Not same function
7
How to Tell Proteins Apart!
- Sequence and fold give overall properties
- Molecular weight
- Solubility
- Exposed hydrophobic surface
- Ability to bind other molecules, metals
- pI- the overall charge of the protein
- Sequence!!!
- To characterize properties, separate the protein
from all other cell contents
8
Protein Purification Techniques
- A. Simple solubility characteristics-
precipitation - Temperature
- pH
- Salting out
- Different proteins precipitate under different
solution conditions- can use soluble or insoluble
fractions
9
Protein Purification Techniques
- B. Chromatography- fractionation of contents
in solution based on selection by a stationary
phase - Size- sieve effect, small molecules faster
- Ion exchange- charge attraction at protein
surface - Choose stationary phase for proteins with
more - charge - First bind everything, then elute with salt
- Hydrophobic interaction- hydrophobic accessible
surface - Affinity chromatography
- Antibody, binding protein
- Inserted tag (e.g. 6-His)
10
Protein Purification Techniques
- C. Gel Electrophoresis- migration in a gel
matrix (size and shape) driven by an electric
field (charge) - Sieving effect
- Relative charge
- Visualization- staining with dye, fluorescent
antibody (Western blotting) - SDS- protein denaturant, enables separation based
almost exclusively on molecular weight - Iso-electric focusing- method to measure pI, but
also can be used for separation
11
Chromatography and SDS-PAGE
M 1 2 3 4 5 6 7 8 9
(Lanes 3, 4)
(Lanes 1, 2)
I
Fusion protein
54.4
36.5
GST
21.5
(Lanes 7, 8, 9)
14.4
T-ag
Volume (ml)
12
Protein Characterization
- A. Sequence
- Amino acid analysis- total digest, then count how
much of each amino acid - Edman stepwise degradation- cleave of one residue
at a time, then identify - Peptide mapping- cleave into fragments, then
identify - Direct sequencing by Mass Spectrometry
- Exact molecular weights
- Characteristic fragmentation
13
Protein Characterization
- B. Spectroscopic properties
- UV-Vis- Backbone, Phe, Tyr, Trp, co-factors
- Infrared/Raman- characteristic bond vibrations
- Circular Dichroism (CD)- backbone conformation
- Fluorescence
- Intrinsic- Trp, Tyr
- Attached dyes- Cys
- Electron Paramagnetic Resonance (EPR)
- Metals, free radicals
- Attached probes
- Nuclear Magnetic Resonance (NMR)
- Many probes viewed simultaneously
- Structure and dynamic processes
14
Protein Characterization
- C. Antibodies
- Use protein of interest to raise antibodies
(rabbit) - Different antibodies can recognize different
regions (epitopes) - Can distinguish differences as small as 1 residue
- Attachment of indicators- dyes, radioactivity
- Applications- e.g. immunoassay, ELISA