Simpleminded molecular homology - PowerPoint PPT Presentation
1 / 45
Title:
Simpleminded molecular homology
Description:
... molecular ... Molecular homology. Two proteins in two different organisms may be encoded ... is of interest to molecular evolutionary biologists ... – PowerPoint PPT presentation
Number of Views:85
Avg rating:3.0/5.0
Title: Simpleminded molecular homology
1
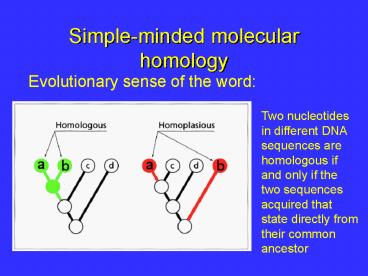
Simple-minded molecular homology
Evolutionary sense of the word
Two nucleotides in different DNA sequences are
homologous if and only if the two sequences
acquired that state directly from their common
ancestor
2
Molecular homology
- Two proteins in two different organisms may be
encoded by the same gene - I.e., the genes are direct descendants of a gene
in the MRCA. They are homologous. - The two genes may share many aa in common and
have similar function - But, if functionality has been acquired
independently, then functionality is not
homologous
3
Classes of molecular homology
- For many, still, similarity homology (read
Reeck et al 1987). Isology? - Most genes are evolutionarily related extant
genomes were derived by duplication,
modification, and recombination of a small number
(one?) of original replicators - Most genes are at some level homologous?
- Need to constrain the term to make it useful
4
Different processes of divergence result in
different classes of homology
- Speciation the divergence of lineages of
organisms - Gene duplication the divergence of lineages of
genes within an organismal lineage - Horizontal gene transfer the divergence of
lineages of genes by transfer across different
organismal lineages
5
Each of these processes is of interest to
molecular evolutionary biologists
- Each process results in genes that trace back to
a single genealogical precursor - But, all three processes are not studied
simultaneously, so we need to make distinctions
when only one of these processes is of interest - Walter Fitch (1970) and Gray and Fitch (1983)
proposed the following terms...
6
Molecular homology
- Orthologous genes diverged as a result of a
speciation event - Paralogous genes diverged as a result of a gene
duplication event - Xenologous genes diverged as a result of lateral
gene transfer
7
Evolution of globin genes in vertebrates
- All genes can be traced back to common ancestor,
so they are all homologous (at some level) - Duplication event gave rise to the a- and b-
hemoglobin gene families - All tetrapods have both types of globin genes
8
To reconstruct organismal phylogeny we need to
compare orthologous genes
9
Concerted evolution
- Above arguments assume that duplicated genes will
evolve independently following the duplication
event - But many duplicated genes will continue to
interact - Observation
- multiple copies of many repeated gene families
are very similar within an individual and within
a species - But they were quite different among closely
related species
10
Concerted evolution
- If duplicated genes evolved independently, expect
greater within species than between species
divergence (among duplicated genes). Left bottom
11
Concerted evolution
- Under concerted evolution, expect greater between
species than within species divergence (among
duplicated genes). Right bottom
12
Six kinds of nucleotide substitution
13
Substitutions in mt COII gene among bovids
14
The need to correct observed sequence differences
15
Possible substitutions among four nucleotides
16
Rate 3a
K 2(3at)
17
Temporal change in the probability of having a
certain nucleotide, say A, at a given nucleotide
site
18
(No Transcript)
19
Using two parameters transitions and
transversions
20
Temporal change in the probability of having a
certain nucleotide, say A, at a given nucleotide
site
21
Jukes-Cantor one parameter model
Kimuras 2 parameter model
22
Jukes-Cantor is a special case of Kimuras
model(they are nested models)
JC
K2P
When alfa beta these two are the same
23
Felsenstein 1981s model
- Base composition may cause some substitutions to
be more frequent than others - If a sequence contains very few Gs, then we would
not expect to see many changes involving that
nucleotide - This model allows the frequencies of the four
nucleotides to be different - Jukes Cantor is a special case of this model too,
when all nucleotides have the same frequency
24
Felsenstein 1981
pi is the frequency of the ith base averaged over
the sequences being compared
If pApCpGpT then F81 JC
25
Hasegawa, Kishino, Yano 1985
- This model merges the K2P and F81 models
- Allows TS and TV to occur at different rates
- Allows base frequencies to vary
- JC, K2P, and F81 call be considered special cases
of this model (they are nested)
26
HKY85
27
General time-reversible model
6 parameters in substitution matrix
28
Any Model
Substitution probability matrix
Base composition vector
29
(No Transcript)
30
More models
31
Real DataObserved and expected changes
- Comparison of human and chimp mtDNA sequences
(307/1333 sites are different) - K2P assigns P0.22, Q0.011
- HKY85 assigns A0.37, T0.18, C0.40, G0.05
- Parameter-rich models more closely approximate
the observed pattern
32
How to choose a model
- More parameters, more realism, but
- Each time we add a parameter, we must estimate
the value for the parameter using the data - The more parameters we add, the greater
uncertainty in our estimates (sampling error
increases) - Fewer parameters inaccurate estimates
- Many parameters low precision
33
Likelihood
- Given observed data and an hypothesis, how can we
decide if the hypothesis is an adequate
explanation? - Probability of observing the data given a
particular model. LPr(DH) - As we have seen, different models may make the
observed data more or less probable
34
Likelihood
- Distinguish between
- Pr (getting observed data)
- Pr (the underlying model being correct)
- Sobers example Loud noise and gremlins playing
bowling. - Likelihoods for different models can be compared
if the models are nested (LRT 2x difference
between log-L is Chi-square distributed)
35
lnL-2064.8
lnL-2691.8
lnL-2424.8
lnL-2075.4
36
Good question
- Why is the likelihood of the observed data not
L1? - Likelihood is the probablity of observing the
data given the model (LPr (DH) - If we could calculate this value for all possible
data sets, the sum would be one - Since we are only concerned with the Pr of one of
those data sets (the observed data), then L lt1
37
More Assumptions
- All nucleotide sites change independently
- The substitution rate is constant over time and
in different lineages - The base composition is at equilibrium
- The conditional probabilities of nucleotide
substitutions are the same for all sites, and do
not change over time - Most of these are not true in many cases
38
Independence
39
Base composition
LogDet transformation recovers additive distances
between sequences even when base composition is
variable
40
Rate variation among sites
- Equal rates among sites simplifies the math, but
at a cost to biological realism - Figure shows different rates of substitution in
different parts of the genome of mammals
41
Invariant sites (I)
- If some sites are constrained to vary by
selection - Sequences that evolve fast may show less
divergence than sequences that are slower
A 0.5 rate, 20I B 2 rate, 50 I
42
Gamma distribution
Allow more than just two categories (zero and
non-zero rates)
43
(No Transcript)
44
How to chose a model? Modeltest
45
How to choose parameters for your model?
- Alternative 1 estimate from data directly as
each tree is being evaluated (time consuming) - Alternative 2 find a reasonable tree and
estimate parameters based on that tree (Modeltest
will do this for you) - Alternative 3 iterate on ML trees obtained with
parameters estimated in step 2, until nothing
changes