Lectures - PowerPoint PPT Presentation
1 / 17
Title: Lectures
1
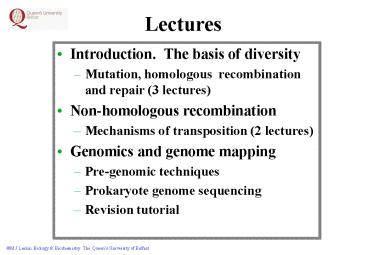
Lectures
- Introduction. The basis of diversity
- Mutation, homologous recombination and repair (3
lectures) - Non-homologous recombination
- Mechanisms of transposition (2 lectures)
- Genomics and genome mapping
- Pre-genomic techniques
- Prokaryote genome sequencing
- Revision tutorial
2
Non-homologous recombinationTransposable elements
LECTURE
- A. Discovery
- B. Classification
- C. Examples and distribution
- Antibiotic resistance spread
- Cassette model
- D. Mechanisms
- E. Regulation
- F. Methods of study and uses
4
5
3
A. Discovery
- First noted in 1967 in E.coli as cause of polar
mutations in - gal operon (Saedler) / lac operon (Shapiro)
- High frequency of spontaneous reversion to gal
or lac - Hedges and Jacob (1974) demonstrated 1st
Transposon Tn1 (Tn3 related) Ampr in plasmid RP4
gal operon on defective lambda phage ?dgal
4
IS1 mediated excisions in the gal operon
galIS1 in this sector reverts as red WT (gal)
colonies on McConkey Agar. Note the high
frequency
gal point (amber) mutation. Supression revertants
at low frequency
?gal (deletion). No revertants possible
5
Deletion of adjacent genes due to activity of IS1
inserted in the gal operon
Chlorate resistant colonies on chlorate/McConkey
agar. A. IS1 has transposed to chlD locus but
still reverts as papillae to gal B. IS1 has
transposed to chlD locus then gal has been
deleted hence no papillae
6
Deletion of adjacent genes due to activity of IS1
inserted in the gal operon
Close-up of colonies with gal papillae
Chlorate resistant colonies on chlorate/McConkey
agar. A. IS1 has transposed to chlD locus but
still reverts as papillae to gal B. IS1 has
transposed to chlD locus then gal has been
deleted hence no papillae
7
B. Classification
- There are four basic types
- TYPE I The Insertion sequences and their
composite elements - TYPE II The Tn3 family of elements
- TYPE II The transposing bacteriophages (e.g. mu
- not covered here) - The conjugative transposons (e.g. Tn916 carrying
tet resistance around a range of host cells in
Enterococcus and other bacteria). Large family
found in these Gram positive bacteria with broad
host range. Carry Integration / excision
determinants and plasmid transfer genes.
INTEGRATE - EXCISE -TRANSFER ON PLASMID (not
covered in detail here).
8
B. Classification Cont...
- Many features in common but with exceptions.
- All transpose as DISCRETE sequences
- ALL have transposase which serves to recognise
ENDS - MUST have precise end recognition EITHER use
terminal inverted repeat sequences OR in some
cases integrate at specific sequences to produce
a consensus sequence for end recognition
9
C. Examples and distribution
- TYPE I IS1, IS2, IS5, IS10
Inverted repeat
General structure
COMPOSITE TRANSPOSONS
10
C. Examples and distribution cont...
- TYPE II The Tn3 like elements. Much BIGGER!
- Many ANTIBIOTIC RESISTANCE DETERMINANTS
11
C. Examples and distribution cont...
- Antibiotic resistance spread
- largely due to transposable elements
- R100 shows cassette model of evolution
12
D. Transposition Mechanisms
CONSERVATIVE VS REPLICATIVE Independent of
RecA
Donor
CONSERVATIVE TRANSPOSITION
REPLICATIVE TRANSPOSITION
13
E. Regulation
All transposons are under negative
regulation Recombinational frequencies down to
around 10-3 to 10-6 In E. coli the growth
temperature greatly affects many transposition
events. Higher frequencies at lower temperatures
(below 37oC) Especially IS1 and Tn3. Basis not
known. EXAMPLES a. Repressor molecule Tn3 b.
Antisense RNA (Tn10) c. Methylation (Tn10 and
many IS elements) d. Transcriptional frameshift
IS1. fusion of two reading frames
14
E. Regulation cont...
a. Tn3 Repressor
?-lactamase
Transposase
Repressor
15
E. Regulation cont...
b/c. Antisense RNA and methylation.
IS10 best studied
pOUT antisense
pIN
GA(me)TC
Transposase
16
F. Methods of study and uses
Various methods used to demonstrate
transposition. 1. Deletion formation (as for
IS1 before) 2. Cointegrate formation (as for
practical 3) 3. Non-replicating plasmids as
delivery vectors 4. Defective phage such as
lambda as a vector
Can be used a a means to TAG genes for mapping
(see practical) Can be used for insertional
mutagenesis (esp Tn5 in Gm negatives)
? Dale Chapter 7.
17
Plasmid cointegration mediated by IS1. See
practical exercise
Crosses of A. E.coli DP990 (pOXKm, pKPG16
(pBR322IS1)) with C600 nalR B. E.coli DP990
(pOXKm, pBR322 (Control)) with C600 nalR
A. 100µl undiluted and plated on Km,Tet and Nal
agar plates. Cointegrates grow
100µl of 10-3 dilution plated on Km, Nal
plates. Indicates the pOXKm transconjugants
B. 100µl undiluted and plated on Km,Tet and Nal
agar plates. No cointegrates detected