Three dimensional structure of DNA - PowerPoint PPT Presentation
1 / 38
Title:
Three dimensional structure of DNA
Description:
ds DNA predominates in vivo ... Model of the 30nm chromatin fiber shown as a solenoid or helix formed by individual nucleosomes ... – PowerPoint PPT presentation
Number of Views:235
Avg rating:3.0/5.0
Title: Three dimensional structure of DNA
1
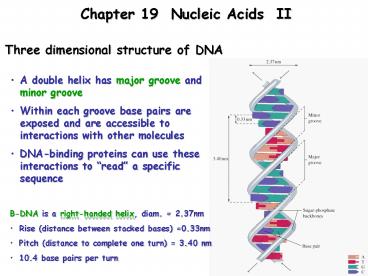
Chapter 19 Nucleic Acids II
Three dimensional structure of DNA
- A double helix has major groove and minor groove
- Within each groove base pairs are exposed and are
accessible to interactions with other molecules - DNA-binding proteins can use these interactions
to read a specific sequence
- B-DNA is a right-handed helix, diam. 2.37nm
- Rise (distance between stacked bases) 0.33nm
- Pitch (distance to complete one turn) 3.40 nm
- 10.4 base pairs per turn
2
DNA binding protein
3
EcoR1 restriction enzyme
Dimerizes and binds one face Makes base contacts
in major groove
4
Weak Forces Stabilize the Double Helix
- Hydrophobic effects. Burying purine and
pyrimidine rings in the double helix interior - (2) Stacking interactions. Stacked base pairs
form van der Waals contacts
(3) Hydrogen bonds. Hydrogen bonding between
base pairs. (4) Charge-charge interactions.
Electrostatic repulsion of negatively charged
phosphate groups is decreased by cations (e.g.
Mg2) and cationic proteins
5
Weak Forces Stabilize the Double Helix
(1) Hydrophobic effects. Burying purine and
pyrimidine rings in the double helix interior (2)
Stacking interactions. Stacked base pairs form
van der Waals contacts
(3) Hydrogen bonds. Hydrogen bonding between
base pairs. (4) Charge-charge interactions.
Electrostatic repulsion of negatively charged
phosphate groups is decreased by cations (e.g.
Mg2) and cationic proteins
ds DNA predominates in vivo
Double-stranded DNA is thermodynamically more
stable than the separated strands
(under physiological conditions)
6
Denaturation of DNA
Denaturation - Complete unwinding and separation
of the 2 strands of DNA .(only in vitro) Heat
or chaotropic agents (e.g. urea) can denature DNA
Local unwinding can occur in vivo
Double-stranded (DS) DNA (pH 7.0), absorbance max
260nm Denatured DNA absorbs 12 -40 more than DS
DNA
Allows easy measurement of DNA melting
7
Heat denaturation of DNA
- Melting point (Tm) - temperature at which 1/2 of
the DNA has become single stranded - Melting curves can be followed at Abs260nm
cooperative
8
DNA Can Be Supercoiled
Overwound or underwound DNA.compensates by
supercoiling to restore 10.4 base pairs per turn
of helix
Over/under winding DNA (stress) can be caused by
various activities
Transcription local unwinding of dsDNA and
helical stress
Replication
9
Topoisomerase
Overwound or underwound DNA.compensates by
supercoiling to restore 10.4 base pairs per turn
of helix
Topoisomerases - enzymes that can alter the
topology of DNA helixes by(1) Cleaving one or
both DNA strands(2) Unwinding or overwinding the
double helix by rotating the strands(3)
Rejoining ends to create (or remove) supercoils
10
Cells Contain Several Kinds of RNA
Four major classes
- Ribosomal RNA (rRNA) - an integral part of
ribosomes, accounts for 80 of RNA in cells - Transfer RNA (tRNA) - carry activated amino acids
to ribosomes for polypeptide synthesis (small
molecules 73-95 nucleotides long)
- Messenger RNA (mRNA) - carry sequence information
to the translation complex - Small RNA - have catalytic activity or associate
with proteins to enhance activity
11
Cells Contain Several Kinds of RNA
1
mRNA
Protein
DNA
4
2
3
RNA
Ribosomal RNA
tRNA and
Non-coding
catalytic
antisense
(Junk DNA)
RNA interference
Guide RNA
Small RNAs
Essential for protein function
12
2009 Nobel Prize in Physiology or Medicine
Elizabeth Blackburn Carol Greider Jack Szostak
Discovery of how chromosome are protected by
telomeres and the enzyme telomerase
TTAGGG
TTAGGG
TTAGGG
TTAGGG
Telomeres
Protects ends of chromosomes
Prevents loss of information during
replications
Added to DNA via enzyme Telomerase
Using RNA template
Aging and cancer
Somatic cells lack telomerase
Limits the of cell divisions ?
13
Telomerase
14
RNA structure
- RNAs are single-stranded molecules
- Often have complex secondary structures
- Can fold to form stable regions of base-paired,
double-stranded RNA (A-form helix) - Example is stem-loop (hairpin) structure
- Stem-loops or hairpins can form from short
regions of complementary base pairs - Stem base-paired nucleotides
- Loop noncomplementary nucleotides
15
tRNA structure
16
DNA Is Packaged in Chromatin in Eukaryotic Cells
How to pack long DNA molecule (genome) into
nucleus
- Chromatin - DNA plus various proteins that
package the DNA in a more compact form - The packing ratio difference between the length
of the metaphase DNA chromosome and the extended
B form of DNA is 8000-fold
Histone-DNA complex nucleosome
Extended Chromatin
Beads-on-a-string
17
Active and silent chromatin
18
Regulation of gene expression
mRNA
Protein
DNA
RNA
Non-coding
Hb
Insulin receptor
amalase
Liver cell RBC Cheek cell
19
Regulation of gene expression
mRNA
Protein
DNA
RNA
Non-coding
Hb
Insulin receptor
amalase
X
X
euchromatin
heterochromatin
Liver cell RBC Cheek cell
20
Nucleosomes
- Histones - the major proteins of chromatin
- Eukaryotes contain five small, basic histone
proteins containing many lysines and arginines
H1, H2A, H2B, H3, and H4 - Positively charged histones bind to
negatively-charged sugar-phosphates of DNA
Histone-DNA complex nucleosome
Extended Chromatin
Beads-on-a-string
21
Nucleosomes
- Histones - the major proteins of chromatin
- Eukaryotes contain five small, basic histone
proteins containing many lysines and arginines
H1, H2A, H2B, H3, and H4 - Positively charged histones bind to
negatively-charged sugar-phosphates of DNA
- Nucleosome beads are DNA-histone complexes on a
string of double-stranded DNA - Each nucleosome is composed of
- Histone H1(1 molecule)
- Histones H2A, H2B, H3, H4 (2 molecules each)
- 200 bp of DNA
22
Diagram of nucleosome structure
- Each nucleosome is composed of
- Histone H1(1 molecule)
- Histones H2A, H2B, H3, H4 (2 molecules each)
- 200 bp of DNA
charge groove
- Packaging of DNA in nucleosomes reduces DNA
length tenfold
23
(No Transcript)
24
30 nm Chromatin Fiber
- DNA is packaged further by coiling of the
beads-on-a-string into a solenoid structure. - Achieves another fourfold reduction in
chromosome length. (4 X 10 40 fold)
- Model of the 30nm chromatin fiber shown as a
solenoid or helix formed by individual
nucleosomes - Nucleosomes associate through contacts between
adjacent histone H1 molecules
25
protein scaffolds in chromatin
Histones have been Removed to visualize scaffolds
Loops attached to scaffold
Protein scaffold
- Chromatin fibers attach to scaffolds
- Holds DNA fibers in large loops
- This accounts for an additional 200-fold
condensation in DNA length. (200 X 40 8000
fold)
26
4x
200x
10x
27
(No Transcript)
28
Regulation of gene expression
mRNA
Protein
DNA
RNA
Non-coding
catalytic
antisense
RNA interference
Regulate mRNA stability RNA processing
29
Nucleases and Hydrolysis of Nucleic Acids
- Nucleases - hydrolyze phosphodiester bonds
RNases (RNA substrates) DNases (DNA substrates) - Exonucleases start at the end of a chain
- Endonucleases hydrolyze sites within a chain
Required for synthesis/repair of DNA
synthesis/degradation of RNA
May cleave either the 3- or the 5- ester bond
of a 3-5 phosphodiester linkage
30
OH group of RNA
OH
Can form H-bonds in RNA mol
Can participate in certain chemical rxns
OH
OH
Diff chemical reactivity than DNA
OH
31
Alkaline Hydrolysis of RNA
Unstable incubation with NaOH
2 OH acts as a catalyst
Intramolecular transesterification
Demonstrates the diff in chem Reactivity of DNA
vrs RNA Due to 2 OH
32
RNase A
Uses three fundamental catalytic mechanisms
Proximity effect position phosphodiester between
2 His residues
Acid-base catalysis (His-119 and His-12)
Transition state stabilized (by Lys-41)
33
(No Transcript)
34
(No Transcript)
35
Restriction Endonucleases
Enzymes that recognize specific DNA sequences and
cut both strands
Substrate for EcoR1
Palindromic seq
Bacteria can restrict the invasion of foreign
(bacteriophage) DNA
protect their own DNA by covalent modification of
bases at the restriction site (e.g. methylation)
Bacteria contain restriction enzymes and
methylases
36
Methylation and restriction at the EcoR1 site
Palindromic seq
Foreign DNA
Unmethylated Substrate for EcoR1
37
Restriction Endonucleases
- Type I - catalyze both the methylation of host
DNA and cleavage of unmethylated DNA at a
specific recognition sequence - Type II - cleave double-stranded DNA only, at or
near an unmethylated recognition sequence - More than 200 type I and type II enzymes are
known - Most recognize palindromic sequences (read the
same in either direction)
38
GGGCCC CCCGGG
GGGCC C
C CCGGG