Web Services for PIR/UniProt Databases - PowerPoint PPT Presentation
1 / 1
Title:
Web Services for PIR/UniProt Databases
Description:
... are designed using Unified Modeling Language (UML) in combination with existing ... Design the system using class diagram in UML. Generate the code ... – PowerPoint PPT presentation
Number of Views:58
Avg rating:3.0/5.0
Title: Web Services for PIR/UniProt Databases
1
Web Services for PIR/UniProt Databases Baris E.
Suzek, Hongzhan Huang, Sehee Chung, Hsing-Kuo
Hua, Peter McGarvey, Zhangzhi Hu, Cathy H. Wu,
Protein Information Resource, Georgetown
University Medical Center, Washington, DC, USA
20057-1455
Abstract Protein Information Resource (PIR) is
an integrated bioinformatics resource that
provides protein databases and analysis tools to
support genomic and proteomic research. PIR
recently joined with the European Bioinformatics
Institute (EBI) and Swiss Institute of
Bioinformatics (SIB) to establish UniProtthe
Universal Protein Resourceto produce a single
worldwide resource of protein sequence and
function, by unifying the PIR, Swiss-Prot, and
TrEMBL database activities (http//www.uniprot.org
). The UniProt Knowledgebase (UniProtKB) provides
the central database of protein sequences with
accurate, consistent, rich sequence and
functional annotation. UniProtKB consists of two
sections Swiss-Prot, containing manually-annotate
d records with information extracted from
literature and curator-evaluated computational
analysis, and TrEMBL, containing computationally
analyzed records that await full manual
annotation. One of the biggest challenges in life
sciences research is the discovery, integration
and exchange of data coming from multiple
research groups. To make the PIR resource widely
accessible to the research community and
application programs, we are adopting an
open-source, common-standard distribution
practice and employing industry-standard J2EE
technology to develop protein object models and
web services. To make the PIR resource
interoperable with other bioinformatics
databases, we are developing controlled
vocabularies and common data elements. The web
services is in the framework of the cancer
Biomedical Informatics Grid (caBIGTM), an
infrastructure connecting individuals and
institutions to enable the sharing of data and
tools for cancer research and developed under the
leadership of National Cancer Institutes Center
for Bioinformatics (NCICB). PIR, as a participant
of caBIGTM, is developing Grid-enablement of
PIR/UniProt Data Source project. The goal of
this project is to demonstrate how the
PIR/UniProt data source can be discovered and
consumed in a grid environment by creating an
object layer and a web service layer for
accessing the data source. The project has an
n-tier architecture. The data layer, supported by
Oracle 9i, stores the UniProtKB data. The data
access layer utilizing Hibernate provides the
mapping between relational database and object
model. The object layer is developed using a
Model Driven Architecture (MDA) approach. The use
cases are developed with input from user
community. The objects and their relations are
designed using Unified Modeling Language (UML) in
combination with existing UniProtKB XML schemas.
An object-XML mapping tool (Castor) has been used
to serialize/deserialize XML data from/to
objects. The web service layer, supported by
Apache Axis, provides language-independent
programmatic access to the objects using SOAP
protocol. The web services will facilitate many
query mechanisms to access PIR/UniProt data
Identifier searches such UniProtKB ID, RefSeq
number String-based searches for fields such as
protein, gene name or keywords Boolean
searches The results are returned in XML and
FASTA format for ease data exchange. To address
the issues of data interoperability, PIR is
participating in development of common data
elements (CDE) as a part of caBIGTM Vocabulary
and Common Data Elements (VCDE) activities. As
members of the NIAID Administrative Resource for
Proteomic Research Centers, the PIR team and the
Virginia Bioinformatics Institute are developing
a cyber infrastructure with a central proteomic
database for the NIAID Proteomic Research
Program. We have established an Interoperability
Working Group (IWG) to discuss and address
database interoperability issues. Interconnecting
with the IWG and caBIG VCDE activities, we also
participate in the HUPO PSI, focusing on mass
spectrometry (PSI-MS) and general proteomics
standards for formats (PSI-ML, XML format for
data exchange), minimum reporting requirements
(MIAPE), and ontologies (PSI-Ont).
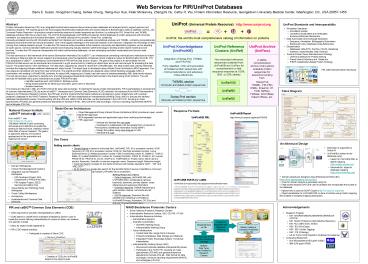
Model Driven Architecture
Response Formats
National Cancer Institute caBIGTM Initiative
- Object Management Groups Model Driven
Architecture (MDA) provides an open,
vendor-independent approach - MDA separates business and application logics
from underlying technologies - PIRs approach
- Analyze and develop the use cases
- Developed in collaboration with the adopter
from University of Pennsylvania, BioMedical
Informatics Facility (BMIF) - Design the system using class diagram in UML
- Generate the code
UniProtKB Report http//www.pir.uniprot.org/entry/
P00439
UniProtKB XML
From caBIGTM site (http//cabig.nci.nih.gov/) V
oluntary network or grid connecting individuals
and institutions to enable the sharing of data
and tools, creating a World Wide Web of cancer
research. The goal is to speed the delivery of
innovative approaches for the prevention and
treatment of cancer
- Use Cases
- Setting search criteria
- Simple Search is based on individual field
UniProtKB, PIR, ID or accession number, NCBI
Taxonomy ID, PIR ID or accession number, NCBI GI,
GenPept accession number, Locus ID/Entrez Gene
ID, Refseq accession number, PDB ID with/without
chain ID, OMIM ID, TIGR ID, EMBL ID,
UniRef100/90/50 ID, UniParc ID, PubMed ID(PMID),
PIRSF ID, PFAM ID, EC number, PROSITE ID, PRINTS
ID, GO ID, InterPro ID, TIGRFAMS ID, Protein
name, Gene name or symbol, Keywords, Scientific
or common organism name, Sequence length,
Molecular weight - Advanced Search is based on two fields combined
with boolean operators AND , OR and AND_NOT
- All-ID Search is a google-like search for the
identifier fields if source of identifier is not
known - Batch Retrieval using multiple UniProtKB IDs or
accessions
Architectural Design
- Data layer is supported by Oracle 9i
- UniProtKB is loaded to the database using
- Castor for UniProtKB XML to object mapping
(http//castor.exolab.org) - Hibernate for object to database mapping
(http//www.hibernate.org)
- Domain Workspaces
- Clinical Trial Management Systems
- Integrative Cancer Research Workspace
- PIR Developer Project Grid Enablement of
PIR/UniProt Data - PIR Adopter Project SEED Genome Annotation Tool
- Tissue Banks and Pathology Tools Workspace
- Cross Cutting Workspaces
- Architecture
- Vocabularies and Common Data Elements
- Setting Response Criteria
- Default response UniProtKB XML with UniProtKB
ID/AC, protein/gene name(s), keywords, taxonomy,
primary citation, cross-references and sequence
information - Extended response Default response plus gene
location, feature, comments and all citations - FASTA response Sequence file with identifier
line containing UniProtKB ID, UniProtKB
Primary_Accession, GO ID(s) and species name and
protein name
- Domain objects are designed using Enterprise
Architect (EA) (http//www.sparxsystems.com/ea.htm
) - Code for domain objects is generated using EA
- Data access objects (DAO) are used to abstract
and encapsulate the access to the database
UniProtKB FASTA for caBIG gtUniProKB ID
AccessionGO ID(s)Organism NameProtein
Name gt1433B_HUMAN P31946GO0005515Homo
Sapiens14-3-3 protein beta/alpha MAQPAELSREENVYMA
KLAEQAERYEEMVEFMEKVAKTVDSEELTVEERNLLSVAYKNVI GARRA
SWRIISSIEQKEESRGNEDRVTLIKDYRGKIEVELTKICDGILKLLDSHL
VPSST APESKVFYLKMKGDYYRYLAEFKSGTERKDAAENTMVAYKAAQE
IALAELPPTHPIRLGL ALNFSVFYYEILNSPDRACDLAKQAFDEAISEL
DSLSEESYKDSTLIMQLLRDNLTLWTS DISEDAAEEMKDAPKGESGDGQ
- Apache Axis is used as SOAP Engine
(http//ws.apache.org/axis/) - Object serialization to UniProtKB XML is done
at runtime using Castor mapping files instead of
complied mapping descriptors
NIAID Biodefense Proteomic Centers
Acknowledgements
PIR and caBIGTM Common Data Elements (CDE)
- Seven National Proteomic Research Centers
- Administrative Resource Centers SSS, GU-PIR,
VT-VBI - Administrative Resource Activities
- Administrative Support
- Scientific Coordination
- Scientific Working Group
- Interoperability Working Group
- Cyber Infrastructure
- Central Web Site Single Point of Access
- Proteomic Database Data Storage and Retrieval
- Integrated Protein Knowledge System Functional
Interpretation - Interoperability Working Group (IWG)
- Discuss and address database interoperability
issues - Participate in the HUPO PSI, focusing on mass
spectrometry (PSI-MS) and general proteomics
standards for formats (PSI-ML, XML format for
data exchange), minimum reporting requirements
(MIAPE), and ontologies (PSI-Ont).
- CDEs required for semantic interoperability in
caBIG - CDEs stored in caDSR which maintains metadata
to permit a user to locate the correct defining
characteristics of a piece of datum, an instance
of a specific concept - UMLs for object model registered to
- PIRs CDE related activities
- Participate in creation of Gene CDE
- Genomic Identifiers
- Taxonomy
- Creation of CDEs for UniProtKB based on the
object model
- Research Projects
- NIH NHGRI/NIGMS/NLM/NIMH/NCRR/NIDCR (UniProt)
- NIH NIAID (Proteomic Administrative Resource)
- NIH NCI caBIG (Grid, SEED)
- NSF BDI (iProClass)
- NSF SEIII (Entity Tagging)
- NSF ITR (Ontology)
- US Air Force EOS (Epidemic Outbreak
Surveillance) - Computing Resources
- Sun Microsystems AEG grant (V880)
- IBM SUR grant (P690)