Translation Regulation - PowerPoint PPT Presentation
1 / 24
Title:
Translation Regulation
Description:
Translation Regulation. Regulation via elements in the 5' UTR: ... In in vitro translation experiments, introduction of RNA spacers between the cap ... – PowerPoint PPT presentation
Number of Views:239
Avg rating:3.0/5.0
Title: Translation Regulation
1
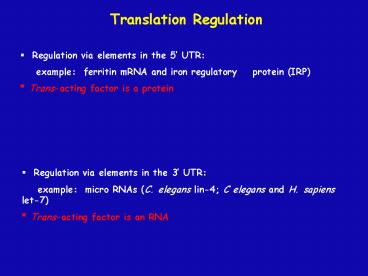
Translation Regulation
- Regulation via elements in the 5 UTR
- example ferritin mRNA and iron regulatory
protein (IRP) - Trans-acting factor is a protein
- Regulation via elements in the 3 UTR
- example micro RNAs (C. elegans lin-4 C
elegans and H. sapiens let-7) - Trans-acting factor is an RNA
2
Translation regulation by 5 UTR sequences
Ferritin mRNA
- Cellular iron homeostasis in mammalian
cells is maintained by the coordinate regulation
of the expression of the transferrin receptor and
ferritin. The amount of iron acquired is
proportional to the number of transferrin
receptors the degree of iron sequestration
within the cell is proportional to the
cytoplasmic level of ferritin.
- Transferrin receptor expression is regulated at
the level of mRNA stability - ? Iron ? ? transferrin receptor mRNA stability
- This leads to increased acquisition of iron
- Ferritin expression is regulated at the level
of translation - ? Iron ? ? translation of ferritin mRNA
- This leads to an increase in available cellular
iron - ? Iron ? ? translation of ferritin mRNA
- This leads to a decrease in available cellular
iron
3
- Sequencing of ferritin genes from a variety of
species revealed the presence of a conserved
element in the 5 UTRs of all ferritin mRNAs. - Deletional anaylsis and construction of
chimeric genes established and RNA element of
about 30 nucleotides as necessary and sufficient
for the iron reposnsiveness of ferritin
translation. - Element is known as iron responsive element or
IRE
Experimentally derived consensus IRE
4
- One copy of the IRE is found in the 5 end of
all known ferritin mRNAs. - Later also found in the 5 UTRs of several
other iron-responsive mRNAs.
5
The IRE is bound by proteins called iron
regulatory proteins (IRP-1 and IRP-2)
High iron No IRP binding Increased ferritin
translation
Low iron IRP binding Decreased ferritin
translation
Increased available cellular iron
Decreased available cellular iron
IRP activity/abundance is regulated by iron
6
- Position effect of the IRE
- IRE is ALWAYS within the first 40 nts of the
mRNA - In in vitro translation experiments,
introduction of RNA spacers between the cap and
the IRE lead to strong reduction of translation
repression. Distance between the IRE and the
initiator AUG was of little consequence.
- Three possible models that accommodate the
position effect - IRE/IRP complex blocks binding of eIF-4F to the
cap - IRE/IRP complex interferes with cap-proximal
binding of the 43S preinitiation complex (40S
subunit, met-tRNA, eIF2) to the mRNA - IRE/IRP complex blocks progression of scanning
43S pre-initiation complex
7
IRE/IRP prevents stable association of the 43S
pre-initiation complex with mRNA
- Experiments with cell-free translation systems
and sucrose gradients showed that the 43S
pre-initiation complex did not associate with the
wild type ferritin 5 UTR in the presence of IRP.
- If the IRE of the ferritin 5 UTR was mutated
so it did not bind IRP, the 43S pre-initiation
complex did bind.
IRP 43S pre-initiation complex association with
mRNA is sterically blocked and there is no
translation
Steric blockage of 43S association
No IRP association of 43S pre-initiation
complex and translation
8
micro RNAs (miRNAs) and short interfering RNAs
(siRNAs)
- Important regulators of eukaryotic mRNAs
- Promote translation repression
- Promote mRNA decay
- 21-26 nt RNAs
- can be functionally equivalent, but are
distinguished - by their mode of biogenesis
(RNAi)
- Originally described in C. elegans now 100s
known - in plants and animals
- Bioinformatics suggests that up to 30 of human
genes - may be regulated by miRNAs
9
miRNA biogenesis
Primary miRNA transcript (pri-miRNA)
- Produced from stem-loop transcripts (pri-mRNA)
- Processed in nucleus by RNase III (Drosha) and
Pasha - Initial produce is 65-75 nt pre-miRNA which is
transported to cytoplasm - Further processed by cytoplasmic RNase III Dicer
complex
Drosha-Pasha complex
Precursor miRNA (pre-miRNA)
Nuclear pore
nucleus
cyto
ATP-dependent assembly with Dicer complex in
cytoplasm
Dicer
Dicer
10
siRNA biogenesis
- Produced from long dsRNA precursors, either
endogenous or exogenously provided - Endogenous siRNAs arise from antisense
transcription or infection by dsRNA viruses - Production is also Dicer-dependent
11
RISC complexes
- RNA-induced silencing complex
- Effector of miRNA and siRNA action
- Final processing of miRNA and siRNA by Dicer
complex - coupled to assembly of RISC complex
ATP-dependent assembly of miRNA into RISC complex
Argonaute (Ago) Family of proteins which are
key components of all RISC complexes
12
How is the function of an siRNA or miRNA
determined?
mRNA degradation Single, perfectly base-paired
RNA (10-11 nt)
Translation inhibition Multiple, imperfectly
base-paired RNAs
13
Different RISC complexes contain different Agos
and perform different functions
- For example
- Drosophila Ago1 miRNA-mediated translation
repression - Drosophila Ago2 siRNA-catalyzed
endonucleolytic cleavage - Interaction of miRNA and Ago with the mRNA may
be influenced by other sequence-specific
RNA binding proteins, providing an additional
level of specificity to miRNA-mRNA interactions
14
(No Transcript)
15
(No Transcript)
16
(No Transcript)
17
lin-4 regulation of lin-14 expression takes
place at the translational level
lin-14 protein is down-regulated, but lin-14 RNA
is not, in a developmentally regulated manner
18
Complementarity between lin-4 RNA and the lin-14
3 UTR is important in regulation of lin-14
protein levels
- Fusion of lin-14 3 UTR to lacZ results in
appropriate temporal regulation of B-gal activity
in C. elegans - Appropriate temporal regulation of this
construct requires lin-4 (i.e., it is not
properly regulated in lin-4 null mutant) - Insertion of 3 copies of the complementary
lin-14 elements into an unrelated 3UTR (lin-14
124 in table below) partially confers temporal
regulation. This indicates that lin-4
complementary sequences in the lin-14 3UTR
mediate part, but not all, of the lin-14
regulation.
19
(No Transcript)
20
(No Transcript)
21
(No Transcript)
22
(No Transcript)
23
(No Transcript)
24
How do miRNAs repress translation?
- Current evidence from experiments with
transfected RNAs in - mammalian cells indicates effect takes place
at translation - initiation and involves the mRNA 5 cap
Full miRNA-mediated translation repression
requires both 5 cap and 3 poly(A) tails