LECTURE 4: Principles of Enzyme Catalysis - PowerPoint PPT Presentation
1 / 16
Title:
LECTURE 4: Principles of Enzyme Catalysis
Description:
An ENZYME is a biomolecular catalyst that ... At time=0, if [P]=0, then. E S ES E P. k-1. k1. Vo = kcat [ES] ... for research and clinical applications ... – PowerPoint PPT presentation
Number of Views:486
Avg rating:3.0/5.0
Title: LECTURE 4: Principles of Enzyme Catalysis
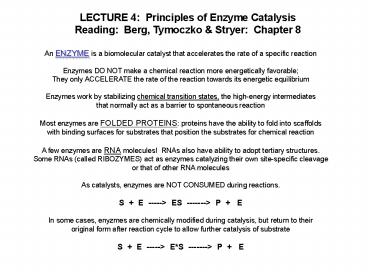
1
LECTURE 4 Principles of Enzyme
Catalysis Reading Berg, Tymoczko Stryer
Chapter 8
An ENZYME is a biomolecular catalyst that
accelerates the rate of a specific
reaction Enzymes DO NOT make a chemical reaction
more energetically favorable They only
ACCELERATE the rate of the reaction towards its
energetic equilibrium Enzymes work by
stabilizing chemical transition states, the
high-energy intermediates that normally act as a
barrier to spontaneous reaction Most enzymes are
FOLDED PROTEINS proteins have the ability to
fold into scaffolds with binding surfaces for
substrates that position the substrates for
chemical reaction A few enzymes are RNA
molecules! RNAs also have ability to adopt
tertiary structures. Some RNAs (called RIBOZYMES)
act as enzymes catalyzing their own site-specific
cleavage or that of other RNA molecules As
catalysts, enzymes are NOT CONSUMED during
reactions. S E -----gt ES -------gt P
E In some cases, enyzmes are chemically modified
during catalysis, but return to their original
form after reaction cycle to allow further
catalysis of substrate S E -----gt ES
-------gt P E
2
Proteases Are Examples of Enzymes That Catalyze
An Energetically Favored Process
Fig 8.0new (peptide hydryolys)
Peptide hydrolysis is an energetically favorable
process, but normally occurs very
slowly. PROTEASES are enzymes that catalyze
peptide hydrolysis. Some proteases are rather
NONSELECTIVE (e.g., papain) Other proteases are
VERY SELECTIVE (e.g., trypsin, thrombin, fibrin)
Fig 8.1new
3
Some Enzymes Employ Cofactors
Some enzymes use cofactors as part of the active
site in enzymatic catalysis APOENZYME
COFACTOR --------gt
HOLOENZYME Many cofactors cannot be synthesized
by humans, and must be obtained through diet as
vitamins and minerals
Tab8.2new
4
Free Energy of Biochemical Reactions
For reaction A B
C D DG is the differential in free
energy between the products vs. reactants If DG
lt 0, reaction is energetically favorable
I.e., reactants will convert to products as
system moves to equilibrium If DG 0, reaction
is already at equilibrium I.e., there
will be no NET conversion of reactants to
products If DG gt 0, reaction is disfavored
I.e., products will convert to reactants as
system moves to equilibrium the
reverse reaction is favored
Free energy DG is usually expressed in units
kcal/mol or kcal x mol-1
5
Standard Free Energy is Related to Equilibrium
Constant
For reaction A B
C D
DG is free energy change when reactant and
product concentrations are A,B,C,D DGo is
free energy change when reactant and product
concentrations are each 1M
WHAT DOES THIS MEAN?
DGo is measure of whether reactants or products
are favored if all components are at same
concentration The actual concentration of
reactants and products impacts on DG even if DGo
is unfavorable, high ratio of reactants to
products can give favorable DG DGo can be
related to the equilibrium constant, Keq, of the
reaction
DG o - RT lnKeq - 2.3RT log10Keq
Keq
Since
6
DG o - RT lnKeq - 2.3RT log10Keq
Tab8.3new
7
Enzymes Accelerate Rate Constant Without Altering
Equilibrium Constant
S
P
Keq kf / kr
Without enzyme
S
P
Keq kf / kr
With enzyme
Fig8.2new
8
Enzymes Stabilize Reaction Transition State(s)
Fig8.3new
9
Properties of Enzyme Active Sites
Active site consists of atoms on residue side
chains that are brought together by the
fold Most of the enzyme structure is a scaffold
to precisely position active site
residues Active site uses range of noncovalent
bonding mechanisms to bind substrate By binding
multiple substrates in a favorable interspatial
relationship and/or by altering charge
distribution (resonance) within substrates,
DGtransition is much smaller than would be
spontaneously
Fig8.7new
Fig8.8new
10
The Michaelis-Menton Model of Enzyme Function
k2
k1
E S ES E P
k-1
k-2
k1
kcat
At time0, if P0, then
E S ES E P
k-1
Vo kcat ES
ES determined by E, S, and rate constants
THESE EQUATIONS CAN BE SOLVED TO EXPRESS THE
REACTION RATE AS A FUNCTION OF THE SUBSTRATE
CONCENTRATION S AND TWO INHERENT PROPERTIES OF
THE ENZYME KM AND kcat
Michaelis-Menton Equation
where
KM k-1 /k1 ES/ES
and
VMAX kcat E
11
The Michaelis-Menton Equation Meaning Behind
The Terms
No matter how large the substrate
concentration, reaction rate can never exceed
VMAX VMAX reflects the TURNOVER RATE of
substrate molecules through the enzyme (kcat) and
the enzyme concentration KM is the substrate
concentration at which reaction rate is HALF
MAXIMAL KM reflects the BINDING AFFINITY of the
enzyme for the substrate The higher the
affinity, the smaller is Km By performing
experiments to calculate Vo at different
substrate concentrations, properties VMAX and Km
can be calculated If the enzyme concentration is
known, VMAX can be used to calculate kcat
Fig8.12new
12
Lineweaver-Burk Plot Facilitates Calculation of
KM and VMAX
Fig8.13new
By inverting equation, get
(
)
1
1
KM
1
Vo
VMAX
VMAX
S
Tab8.5new
13
Many Enzymatic Reactions Proceed Through Fixed
Sequential Steps
PyrLac.p223new
TransAm.p224new
Reaction Intermediate May Utilize Covalently
Modified Enzyme or Cofactor
14
Enzyme Inhibition
Many small molecules can bind to enzymes and
inhibit them. Inhibitors can be described as
REVERSIBLE or IRREVERSIBLE. Inhibitors may be
naturally occuring within the homologous organism
or in a heterologous organism Other inhibitors
are synthetic and have been developed as
pharmaceuticals for research and clinical
applications COMPETITIVE INHIBITORS act by
occupying the enzyme active site in place of the
substrate NONCOMPETITIVE INHIBITORS bind away
from the active site, but their binding exerts
allosteric effects that prevents bound substrate
conversion to product
Fig8.15Anew
Fig8.15Bnew
Fig8.15Dnew
15
Kinetics of Competitive Inhibition
S
Vo VMAX
S KM
Ki reflects the AFFINITY OF INHIBITOR for the
enzyme Inhibitor in effect raises the apparent
KM term The potency of inhibitor determined by
Ki Therefore, the amount of substrate needed for
half-maximum rate is increased THERE IS NO
EFFECT ON VMAX I.e., a competitive inhibitor can
be overcome by sufficiently high substrate
concentration
Fig8.17new
16
Kinetics of Noncompetitive Inhibition
As before, Ki reflects the AFFINITY OF
INHIBITOR for the enzyme Inhibitor reduces VMAX
of the enzyme The potency of inhibitor
determined by Ki The inhibitor does not affect
KM , i.e., the binding of substrate to enzyme
Fig8.19new


























![Welcome to CHEM BIO 3OA3! Bio-organic Chemistry [OLD CHEM 3FF3] PowerPoint PPT Presentation](https://s3.amazonaws.com/images.powershow.com/P1258891208FGTDt.th0.jpg?_=20140110098)



