Gene Prediction - PowerPoint PPT Presentation
1 / 22
Title:
Gene Prediction
Description:
It is a CaM Kinase II protein. Protein Link. exon. intron ... Hidden States ... Probabilistic Hidden State Inference and Model 'Training' The two key problems ... – PowerPoint PPT presentation
Number of Views:137
Avg rating:3.0/5.0
Title: Gene Prediction
1
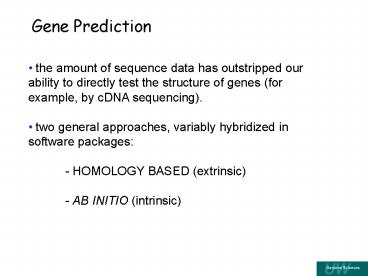
Gene Prediction
- the amount of sequence data has outstripped our
ability to directly test the structure of genes
(for example, by cDNA sequencing). - two general approaches, variably hybridized in
software packages - - HOMOLOGY BASED (extrinsic)
- - AB INITIO (intrinsic)
2
Homology Based Approach (hand version)
- use evolutionary divergence as a tool.
- based on the principal that introns and other
non-coding regions usually diverge much faster
over time. - helped by the fact that many splice patterns
have been determined experimentally (cDNA). - fails when the divergence of distant sequences
is too extreme (rapidly evolving proteins or
large evolutionary distances).
3
Homology Based Approach (cont.)
- for a segment to predict use blastx search to
find closest match in known protein sequences.
blastx translates the query in all frames and
searches a protein database - possibly validate the matched sequence it
might be predicted too! - use that match (or several best matches) as
query in a tblastn search of the segment to
predict. tblastn translates a nucleotide
database in all frames and searches it with a
protein query - locate good exon matches and use as guide to
find splice sites etc.
(note there are automated programs that do this
and some that also try to integrate this with ab
initio prediction)
4
BLASTX of arthropods with first 5 Kb of cosmid
K11E8 from C. elegans
5
Protein Link
Link to Drosophila protein sequence that was high
on the blastx hit list. It is a CaM Kinase II
protein.
6
Excerpt from tblastn search of C. elegans genome
with Drosophila CaMKII sequence
Finally, incorporate a quantitative description
of splice junctions to find best exon-intron
boundaries, guided by homology blocks.
7
Automated software for homology-based prediction
- Genewise (uses predicted proteins from one
genome to drive exon finding in a second genome,
combined with splice-site model to give full
prediction) - Procustes (similar to Genewise)
- Projector (similar to Genewise, but includes
intron position conservation) - Others (use combined ab initio and DNA
alignment) - Twinscan (builds two genome predictions
together, using conserved DNA signal as one
criterion)
There are links to most of these on the course
web pages.
8
Problems with homology-based prediction
- depends on strong homology match elsewhere (50
to 70 of genes now and improving). - incomplete without addition of precise splice
prediction (which requires a guided ab initio
approach). - main methods (Genewise etc) depend on the query
protein being correct (potentially circular
logic).
9
ab initio approach the purist at work
- start with pure DNA sequence.
- apply information about promoter, splicing, and
terminator rules. - this is (essentially) the way the cell does it,
so it must be possible. - BUT it doesnt work so well (rules complex and
not understood?). - add open reading frame analysis and it works a
lot better. - does NOT use sequence similarity to other known
genes.
10
Where the hell is the coding gene!?
TTTTAGGAATCTTAAGACGTTTTAGCAAAGTTCCAAAATTTCTGAAAAAT
ATTTTTTTTTGGTCGACTTCCAAAATTATGAGTGGCAAAAAATAATTAAT
TGTCATTTTTTGACAATAAATAAAAAATTTTCAAACATTTTTTTGAATTG
TTTTATTATGATATTCGGTCATTTTGGCACCATATTAGTCGTTTTTAACA
CTTTCCCCACTGGCGCTACTCCACCTTTAATATAATTTTTGGATTCAGGG
CCTAACTTTTCAAGTTATCTTACCACTTGTCTGCTATGTTCCTGTTACAT
TTATGTATTGTTGGAATAAGTATTCCGGTAAGGAAATTCATCAATGACCA
AATGTAATTGTTTAAAAAAAATTCAAAATTAAATAATTTTTTTAAAAATA
TTTCCCAAATCAAAGACACGACCGAATTTAATTTGAATTCCCGCGCAAAT
GAGTGACGTCATTTTCGATATTTTCGCGGCCAAATTCTTTGGGTTTTCAT
TATTTTTTCTTCTATATGCGATTTAAAACACCGTTTGCCAATTTTTCAGG
CACTTTAAAATTTCAAAATTGGCCTAAAAACGACAATTAAAAAAATAACG
ACAGAACTGAAAATGCAAAAATATCGAAAATGACGTCACTCAAATTTAAT
TCGGCTGTTTTTTTGATTTTTGAAAAATGAAAAAATCATAAAAAATTTAG
AAAATTTTTCAAAATTTTTTTACAGTCATATTCGGCCATTAGGGTCTATT
TTCTGTCATTTAAAACAAACAAATTGAGCCTACTCCACCTTTAAATAAAT
TTTCCAGGCGACCAAATCCTTATATCACAATACACTCTGGCATTTATGGG
TACACTTCCGTGTATTTTCGATCCGCTACTTCAAATGTATTTTATACTGC
CTTACCGAAAGACGGTTCAGAAATTTATTGCACGTTCTCCATTTAAACCA
AATAACGCCCACGCTGTCAGCTCGATTCCTCGACGTTCTTTTTTGACCCC
CTGAAATTATTTAACTGCGAAAAAAGAAACACTTTGCCTACCTGTTTTGC
CGTACATATTTCTCAGAAAAACAATCGAAAAAAAATGTTCTAGATACATC
AAAAAAGTTAACGGTATTTATATGTACCGGAAATTGGTTAGCACCTCTGG
CAAAATTTGGCAGATCACATCTGCCAAGACACCCACATACCCGAATCATT
GTGTTCCTTTTATACAAGCCATAAAATGCAGCTCAACGATGCAATATTTT
TGAACGCTTCTGTAATTTGTGCGATTTTTATAAATTTTGTCCTGATACTT
TTGATACTCAAAAAATCTCCAGCATCTCTTGGAGCCTATAAGTACCTCAT
GATGTATATCAACATTTTCGAGCTGACCTATGCGATTTTGTATTTTGCGG
AGAAACCGGTGAGTTTTTTTAAAAATAGAATAAAATAGATTAAATTGAAA
ATTTGCAAAACTGGAATGCGATAAAATTTAAAATTTTTTGGAAATAGGCA
GTTTCGTTTTATTGAAAACTCTGACACCCTGAAATTTTGGCAATTGCCAG
ATTTTTCGGAGTTGCAGCCGATTCCGGCAATTACAAAAAAACTTCCAATA
TTCGCCAATTTCAAATTTTGCCAGTTTCTGACATTTCCGGCAACCGTGTA
AATTTGCTGGGTTTCAGACTTTTTGCCAATCGGCAGTTGTCGGATTTTAA
AAATTCTGGAGCTTTTGGCAATTACCAAAAATTGAAAAATTCGGCAATCG
GCAATTTTGCCAGATTTTGGAAACTCTAGAAATTTCTGCAATTACCGAAA
ATGTGAATTTTTATACATTGTTAGTATGTGCGTTTTTCCAATTTCAAATT
TCTGGTATTTTTTGGCAATTAGGAAAATAATAAATTTCCACAATTTCCGA
TTTTCAGAAATTCCGGCAATTATTGAAAATTAAATTTTCCGGCAATCGAC
AATTTTGCCGGATTTTGGAAACTCTATACTTTTTACAATTACCGTAAATA
TGAATTTTTATACATTGTTAATTTTGACGAATTTCCAATTTTAAATTCTT
TATATTTTGGGTAATTACCAAAAGTACATATAAATTCCCACATTTTTTGG
TTTTCAGAAGTTCCAAGTACTGCAATTTTGGCAATTTTTGACATTTTTGG
CAACTAGCAATATCCGCCATTCGGAATGCTTTAAATTTCACGCAGCCAGT
AAATTTTTGGCACTTTTGGAAAATTCGAAATACTCTGATAATTGGCAATT
TTTTTCGTAAATACAGAAAATGAATAAATTCGACAATTTTCATTTTGGCT
AATAAAAGAATGATACGTTTTTTTTTAAGATTATGCTCACAAAAGAGTCT
GCATTTTTAATAATAATGAACTGGAGAGCATCAATATTTCCGAAATATGT
TGCTTGCACTCTAAATCGTAAGTTTTCAAAAATAATTTTTTTAGCCCGCC
AATTTTAACTCTTACAGTGCTTTTCATTGGCTTCTTCGGTATGTCAGTTG
CTATTCTGGCCCTTCATTTCATTTATCGATACCTCAGTATCACAAAGTAA
GCTCAAATATTCGGAGCATTTAAGTGTACCAATTTTTCAGGAGCAATCTA
CTGAAAACATTCGATTCGTCGAAAATTGTGCCGTGGTTTATGATACCATT
GCTGAACGGAATAACGTTTATGTGTACAGCGGGATTTTTAATGCGAGCCG
ACGAGCAAACTGATAGATTTATAAAGTAAGAGCTTCTACTTCATAGCGTG
GTTCTAAAAGTTTCAAAACAGTTTTGATGCACAAAAACTGTAAAAATTAC
AACAAAAAAACAAATATGAATTTTCAATGCTAATTGAGAATTTTCATCTT
TTAAAAAGAAATCTGGAAATTGTGTGAAATTTTTTTTTTTAATTTAAGCA
ATTTGTGAATGAATAAAAATTGTCCAAAAGGCTTTCAGAATGTACATACT
AAAGTATTAAAAGAAGGACTTTTATGGCTTAGAGGAACTGTAAAAAAATA
ACTGTTAAAAATTTGTAGAATTTTTATAAATTTTCATAAAATATTTGTTT
TCCAATTTTGCAACACAATTTTTTTTCGAGGTTTTTGGAAACACTGGTTC
TGACATATTTCTGAAAAAAGTTCGTAAAAACGTTAAATTACATTTTCATA
GGTCAGTATTCCCCGGAAATTTTGAAAAAAAAACCTATAATTAAACAAAA
CAGTTTTTACATTTGTGCTATTTTTCGGTAAATTTTTCACAAAAATTTTG
AGGCCCGAAGAGTTACTTTTTTACTAACTTAGATATTGCTGAATACAAAA
GTTTTTTTGAAAAAATTGTGTGGTAATAACGTAGCATTCGGAATGAAAAA
GCCCAAGCGAGCGAGCCTAAGCCTAATTCTAACCTCATAAAAAGTTACAA
GAAGGTTTTTCCTTGCGCTTGGAGCGCAAAAGAAAATAAAAAGGGCTATT
TAGAGTTAGGAACAAACCCAATTTGAATAAAACATTGGAAATCCCAATCC
AGCAAGCCTAAGGGCCCGAAAAACATACTAGGATGCCCAACTGGAATAAA
ATATAGGAAATCCTTATGACACACCGGCGGTATGGCGCGGCTTAAGCCTA
AATAGCCACTTTTATCAAAATACATTTGAGCGAGGCGGTTGTAAACTATT
CGTTCATTAACAAAAAAAAAAATTTTAAGAAGCAAAAAAAGAGACTATAT
TTAAATTTAAAAATAAATATCATATGTTATCACACCTTACAATTAGAATA
TCACGCCTTAATTTAGATCATCGGAATTAAATATCATCAGAGCTAACGCT
CGCCACTGACGCCAAGCCGTAACCTGAGCCTTAGAATAAGCTTAAGCCTA
AGCCTAAGCCTAGTCCAACGCCTAGGCCTAGGCATAAGCCTAAGTCTAAG
CTGAAGCCTAAGCCTAAGCCTATGCCTAACTAAAATTATAACCGTAAAAA
ATACCTGTTAAAAAATTTATGAATTTATATAAATTTTCTAAAAATTTTTT
TTTCGTTTTACAATATTCAATTTTTCGATGTTCCCTGAAATATCGAATTT
TCAGTGAAAACTATCCACCGCTTGTCAAAAACCTCTCAACTATCAATGAT
CTCTACTATGTGGGCCCATTGTTCTGGCCCAAGTACGCCAACTCAACAAC
CGACCACTTTTTCAGTTGGAAGGCTGCAAGGTTATGCCTGATTGCGATGG
GCTTAATTGTGGGTTCAAGATTAGGCTTAAGCTCTAAACAAATCTTTGCT
CCAGGGATTTTCCACTTCAATAATGGTGTTCTTCGGTCTGAAAGCATATT
TGGTAATGAAAAACTTGATGTCACAGTCAACTTCTTGTGACAAGTTCAAA
AGCATTCAGCAGCAATTACTACTTGCTCTAATTCTGCAAACTTCGATTCC
AGTCCTCTTGATGCATATTTCTGCAACCGCGATTTACCTGACAATATTTT
TGGGAAATTCTAACGAGATTATAGGAGAAACCATTGGATTGACAATTGCA
TTGTATCCCGCTCTGAATCCAATTCCAACAATTTTGATCGTCAAAAATTA
TCGGACTGTGTTGATCAGTGAGTTAAAAAATTTTTTTTTTTTTTAATTAT
CCACTTTTGCCAATTTTTGAAAAATCTATAGCACTGTCGCATGTTCAAAT
CTTTATTGGCAATTTGTCGGTCTGCCGATTTGCCGGAAAATTTCAAATCC
GGTAATTTGCCGATTTGCCGGGAAATTTCGATTCCGGCAACTTGCCGATT
TGCCGGAAAATTTAAATTCCGGCAATTTGCCGATTTGCCGGAAAATTTTA
ATTCCGGCAATTCGCCGATTTGCCGGAAAAATCGTTTGCCGCCCACACAT
GATTTGAACATTAGTGCTTGGAACATTATTCGGACAGGGATTAGCGGCAA
TTGCCGTTCGGCAATTTTTTTTTCGACAAATTCGGCAAATTGCCAATTTT
CATTTCCGGCAATTTACCGATTTGCCGGAACTGTTTAGAGTGATTTTTTA
TACGACGGAAGCACTTAAAACAGCGCATTTTCCCATTTTTTCCAGGTTTC
TTTAGATATTTTCATATAGTTTGCTTACTTTTCAAAATAGATGTAGGAAC
ATTCATAGGATGCGTTCAATTTTGCCGAGATGAATTGCAATTCTGAAATT
TCCAAAAAAGGTGCAAAACCACTATTTGCCGAAAATTTTCGGCAATTGCC
GTGTTTCCGGCAAATTCAGCAAAATCGTCAATTTGCCGATTTGCCGATTT
GCCGGAAATGTTAAATTCCGGCAACTTGCCGATTTGCCGATTTGCCGGAA
ATGCTACTCCACCCTTAAAGATTTTTAACCTGTCATCCCAAATTAACGCC
GGTTTTTCAGATATACTGGCTTATGTCAAACGTCGAGTATTCCGACAAAC
TGCGGTGACTCCACTTGTGTTGGCGGATGTAACAACAATCGCTATGCAAA
ATTTGGCCCCGAACTAGCATTTTCCCATATTTTTGTATTTGAAGGTGGTG
TAGTCTAACTTTTTATTGCGTTATTAGACTCAAAATTGTCTGAAAACACC
GAAGTTCATAATGAAACTTCTTGAAAATTTTTCAAAAAAAAGTTATGACG
GCTCAAAAAATGAGCTAAAATTAGTTACAAATTCAAATTTGACATGTCAG
CGGGTGGAAACTAATTTTTTTGAAATCACCGTCTAATTTTAGGGTTTTCA
ACTCTACTTAGATATTCTAAAGTTGATGGACAAAGCTTTTTTTTAAATGT
TGATTTAAAAAAAAACAAAAAAAAATTCCAGCCGTTGCGACCTTGACAAG
TCGGCCAAATTTCAAATTTTAACTAATTTTTGGCCATTTTTTTAACCCGT
CATAACTATTTTTTGAAAAGTTTTCAAGAAGTTTCATTATGAAATTCGGT
GTTTTCAGACAATTTTGGGT
11
Theres the coding gene! (maybe)
ttttaggaatcttaagacgttttagcaaagttccaaaatttctgaaaaat
atttttttttggtcgacttccaaaattatgagtggcaaaaaataattaat
tgtcattttttgacaataaataaaaaattttcaaacatttttttgaattg
ttttattatgatattcggtcattttggcaccatattagtcgtttttaaca
ctttccccactggcgctactccacctttaatataatttttggattcaggg
cctaacttttcaagttatcttaccacttgtctgctatgttcctgttacat
ttatgtattgttggaataagtattccggtaaggaaattcatcaatgacca
aatgtaattgtttaaaaaaaattcaaaattaaataatttttttaaaaata
tttcccaaatcaaagacacgaccgaatttaatttgaattcccgcgcaaat
gagtgacgtcattttcgatattttcgcggccaaattctttgggttttcat
tattttttcttctatatgcgatttaaaacaccgtttgccaatttttcagg
cactttaaaatttcaaaattggcctaaaaacgacaattaaaaaaataacg
acagaactgaaaatgcaaaaatatcgaaaatgacgtcactcaaatttaat
tcggctgtttttttgatttttgaaaaatgaaaaaatcataaaaaatttag
aaaatttttcaaaatttttttacagtcatattcggccattagggtctatt
ttctgtcatttaaaacaaacaaattgagcctactccacctttaaataaat
tttccaggcgaccaaatccttatatcacaatacactctggcatttatggg
tacacttccgtgtattttcgatccgctacttcaaatgtattttatactgc
cttaccgaaagacggttcagaaatttattgcacgttctccatttaaacca
aataacgcccacgctgtcagctcgattcctcgacgttcttttttgacccc
ctgaaattatttaactgcgaaaaaagaaacactttgcctacctgttttgc
cgtacatatttctcagaaaaacaatcgaaaaaaaatgttctagatacatc
aaaaaagttaacggtatttatatgtaccggaaattggttagcacctctgg
caaaatttggcagatcacatctgccaagacacccacatacccgaatcatt
gtgttccttttatacaagccataaaATGCAGCTCAACGATGCAATATTTT
TGAACGCTTCTGTAATTTGTGCGATTTTTATAAATTTTGTCCTGATACTT
TTGATACTCAAAAAATCTCCAGCATCTCTTGGAGCCTATAAGTACCTCAT
GATGTATATCAACATTTTCGAGCTGACCTATGCGATTTTGTATTTTGCGG
AGAAACCGgtgagtttttttaaaaatagaataaaatagattaaattgaaa
atttgcaaaactggaatgcgataaaatttaaaattttttggaaataggca
gtttcgttttattgaaaactctgacaccctgaaattttggcaattgccag
atttttcggagttgcagccgattccggcaattacaaaaaaacttccaata
ttcgccaatttcaaattttgccagtttctgacatttccggcaaccgtgta
aatttgctgggtttcagactttttgccaatcggcagttgtcggattttaa
aaattctggagcttttggcaattaccaaaaattgaaaaattcggcaatcg
gcaattttgccagattttggaaactctagaaatttctgcaattaccgaaa
atgtgaatttttatacattgttagtatgtgcgtttttccaatttcaaatt
tctggtattttttggcaattaggaaaataataaatttccacaatttccga
ttttcagaaattccggcaattattgaaaattaaattttccggcaatcgac
aattttgccggattttggaaactctatactttttacaattaccgtaaata
tgaatttttatacattgttaattttgacgaatttccaattttaaattctt
tatattttgggtaattaccaaaagtacatataaattcccacattttttgg
ttttcagaagttccaagtactgcaattttggcaatttttgacatttttgg
caactagcaatatccgccattcggaatgctttaaatttcacgcagccagt
aaatttttggcacttttggaaaattcgaaatactctgataattggcaatt
tttttcgtaaatacagaaaatgaataaattcgacaattttcattttggct
aataaaagaatgatacgtttttttttaagATTATGCTCACAAAAGAGTCT
GCATTTTTAATAATAATGAACTGGAGAGCATCAATATTTCCGAAATATGT
TGCTTGCACTCTAAATCgtaagttttcaaaaataatttttttagcccgcc
aattttaactcttacagTGCTTTTCATTGGCTTCTTCGGTATGTCAGTTG
CTATTCTGGCCCTTCATTTCATTTATCGATACCTCAGTATCACAAAgtaa
gctcaaatattcggagcatttaagtgtaccaatttttcagGAGCAATCTA
CTGAAAACATTCGATTCGTCGAAAATTGTGCCGTGGTTTATGATACCATT
GCTGAACGGAATAACGTTTATGTGTACAGCGGGATTTTTAATGCGAGCCG
ACGAGCAAACTGATAGATTTATAAAgtaagagcttctacttcatagcgtg
gttctaaaagtttcaaaacagttttgatgcacaaaaactgtaaaaattac
aacaaaaaaacaaatatgaattttcaatgctaattgagaattttcatctt
ttaaaaagaaatctggaaattgtgtgaaatttttttttttaatttaagca
atttgtgaatgaataaaaattgtccaaaaggctttcagaatgtacatact
aaagtattaaaagaaggacttttatggcttagaggaactgtaaaaaaata
actgttaaaaatttgtagaatttttataaattttcataaaatatttgttt
tccaattttgcaacacaattttttttcgaggtttttggaaacactggttc
tgacatatttctgaaaaaagttcgtaaaaacgttaaattacattttcata
ggtcagtattccccggaaattttgaaaaaaaaacctataattaaacaaaa
cagtttttacatttgtgctatttttcggtaaatttttcacaaaaattttg
aggcccgaagagttacttttttactaacttagatattgctgaatacaaaa
gtttttttgaaaaaattgtgtggtaataacgtagcattcggaatgaaaaa
gcccaagcgagcgagcctaagcctaattctaacctcataaaaagttacaa
gaaggtttttccttgcgcttggagcgcaaaagaaaataaaaagggctatt
tagagttaggaacaaacccaatttgaataaaacattggaaatcccaatcc
agcaagcctaagggcccgaaaaacatactaggatgcccaactggaataaa
atataggaaatccttatgacacaccggcggtatggcgcggcttaagccta
aatagccacttttatcaaaatacatttgagcgaggcggttgtaaactatt
cgttcattaacaaaaaaaaaaattttaagaagcaaaaaaagagactatat
ttaaatttaaaaataaatatcatatgttatcacaccttacaattagaata
tcacgccttaatttagatcatcggaattaaatatcatcagagctaacgct
cgccactgacgccaagccgtaacctgagccttagaataagcttaagccta
agcctaagcctagtccaacgcctaggcctaggcataagcctaagtctaag
ctgaagcctaagcctaagcctatgcctaactaaaattataaccgtaaaaa
atacctgttaaaaaatttatgaatttatataaattttctaaaaatttttt
tttcgttttacaatattcaatttttcgatgttccctgaaatatcgaattt
tcagTGAAAACTATCCACCGCTTGTCAAAAACCTCTCAACTATCAATGAT
CTCTACTATGTGGGCCCATTGTTCTGGCCCAAGTACGCCAACTCAACAAC
CGACCACTTTTTCAGTTGGAAGGCTGCAAGGTTATGCCTGATTGCGATGG
GCTTAATTgtgggttcaagattaggcttaagctctaaacaaatctttgct
ccagGGATTTTCCACTTCAATAATGGTGTTCTTCGGTCTGAAAGCATATT
TGGTAATGAAAAACTTGATGTCACAGTCAACTTCTTGTGACAAGTTCAAA
AGCATTCAGCAGCAATTACTACTTGCTCTAATTCTGCAAACTTCGATTCC
AGTCCTCTTGATGCATATTTCTGCAACCGCGATTTACCTGACAATATTTT
TGGGAAATTCTAACGAGATTATAGGAGAAACCATTGGATTGACAATTGCA
TTGTATCCCGCTCTGAATCCAATTCCAACAATTTTGATCGTCAAAAATTA
TCGGACTGTGTTGATCAgtgagttaaaaaattttttttttttttaattat
ccacttttgccaatttttgaaaaatctatagcactgtcgcatgttcaaat
ctttattggcaatttgtcggtctgccgatttgccggaaaatttcaaatcc
ggtaatttgccgatttgccgggaaatttcgattccggcaacttgccgatt
tgccggaaaatttaaattccggcaatttgccgatttgccggaaaatttta
attccggcaattcgccgatttgccggaaaaatcgtttgccgcccacacat
gatttgaacattagtgcttggaacattattcggacagggattagcggcaa
ttgccgttcggcaatttttttttcgacaaattcggcaaattgccaatttt
catttccggcaatttaccgatttgccggaactgtttagagtgatttttta
tacgacggaagcacttaaaacagcgcattttcccattttttccaggtttc
tttagatattttcatatagtttgcttacttttcaaaatagatgtaggaac
attcataggatgcgttcaattttgccgagatgaattgcaattctgaaatt
tccaaaaaaggtgcaaaaccactatttgccgaaaattttcggcaattgcc
gtgtttccggcaaattcagcaaaatcgtcaatttgccgatttgccgattt
gccggaaatgttaaattccggcaacttgccgatttgccgatttgccggaa
atgctactccacccttaaagatttttaacctgtcatcccaaattaacgcc
ggtttttcagATATACTGGCTTATGTCAAACGTCGAGTATTCCGACAAAC
TGCGGTGACTCCACTTGTGTTGGCGGATGTAACAACAATCGCTATGCAAA
ATTTGGCCCCGAACTAGcattttcccatatttttgtatttgaaggtggtg
tagtctaactttttattgcgttattagactcaaaattgtctgaaaacacc
gaagttcataatgaaacttcttgaaaatttttcaaaaaaaagttatgacg
gctcaaaaaatgagctaaaattagttacaaattcaaatttgacatgtcag
cgggtggaaactaatttttttgaaatcaccgtctaattttagggttttca
actctacttagatattctaaagttgatggacaaagcttttttttaaatgt
tgatttaaaaaaaaacaaaaaaaaattccagccgttgcgaccttgacaag
tcggccaaatttcaaattttaactaatttttggccatttttttaacccgt
cataactattttttgaaaagttttcaagaagtttcattatgaaattcggt
gttttcagacaattttgggt
12
Digression into Markov chains
- Many ab initio gene prediction methods (and
sequence alignment methods by the way), are based
on a probability model called a Markov chain. - Ill digress to describe Markov chains and the
related Hidden Markov Model (HMM), then integrate
them with gene finding.
13
Markov Chains - States
- A first-order Markov chain is a linear series of
states, in which the current state depends only
on the previous state in the chain. - note a second-order chain depends on the
previous 2 states, etc.
(if currently in state A, the next state is state
A 90 of the time and state B 10 of the time,
etc.)
14
Markov Chains Emissions
- In most biological applications, each state
defines one or more emissions, each of which
occurs with a some probability. - For simple DNA sequence modeling, the possible
emissions will be A, C, G, and T. - More generally, for N possible emissions in a
given state, the sum of their probabilities must
be 1 (each step along the chain always emits
something).
15
Sequence Probability in Markov Chains
In the simplest form, there are 4 states, each of
which emits one of the four nucleotides (with
probability 1)
The probability of a sequence x of length L
residues is
or
where
In words, the probability of the entire sequence
is the product of the probabilities that each
state in the chain matches the nucleotide, given
that the previous state matched the previous
nucleotide, given that etc.
(of course, most useful Markov chains will have
more than one emission from each state)
16
Hidden States
- If we have only the emissions from a Markov
chain but not the underlying states, the states
are called hidden. Since DATA represent the
emissions, this is the usual situation. - A simple example comes stretches of GC-rich and
AT-rich DNA. - The Markov chain describing this would have two
states - 1) emits G or C with high probability
- 2) emits A or T with high probability
- - both states have a high probability of staying
in the same state at each step along the chain
(but switch occasionally).
17
Probabilistic Hidden State Inference and Model
Training
- The two key problems in HMM use are
- Getting an accurate model to begin with
- - Usually done by guessing plausible state types,
then training the probabilities on a set of
known state data. - Using the model to obtain a probabilistic
interpretation of a sequence (or other data set). - - Various algorithms available that permit
finding the best state path, the overall
likelihood of all state paths (given the data)
etc.
18
Hidden Markov Model in Gene Prediction (flavor)
- Choose a set of appropriate states (exon,
intron, terminator, etc.). - Choose allowed paths between the states
(transitions). - For each state choose various emission patterns
(for this discussion think of these as things
like an open reading frame of length X). - Select a set of data where the states are known
(experimentally described genes) as a training
set. - Determine the set of transition and emission
probabilities that best match the training set. - Apply the trained model to new sequence and find
the concrete sequence states that match the model
best (most probably).
19
Gene Prediction HMM States
taken from Stormo lab paper
20
A very simple HMM for a two-symbol sequence
emit A or T
2 states A-rich and T-rich
transition probabilities
- a training set with random A and T residues
could produce model parameters where PA and PT
were equal (in both states) and PAT and PTA
could be anything. Question what other
parameters fit? - a training set with stretches of A-rich sequence
interspersed with T-rich sequences would produce
what sort of model parameters?
21
Problems with ab initio prediction
- accuracy varies a lot with complexity of
splicing rules (especially bad in mammals,
especially good in bacteria and yeast, in between
in nematodes). - requires a good species-specific training set (a
set of experimentally known gene structures).
22
Assignment for next Tues.
emissions A or T
emission probabilities
states A-rich and T-rich
- A training set with stretches of A-rich sequence
interspersed with T-rich sequences would produce
what sort of model parameters? (qualitatively) - If the training set changed to include A-rich
sequences in longer blocks (on average), how
would this change the 6 probability values?
(qualitatively up or down) - If the T-rich sequences were much more T-rich,
how would this affect the 6 probability values?
(qualitatively)