Application - PowerPoint PPT Presentation
1 / 1
Title:
Application
Description:
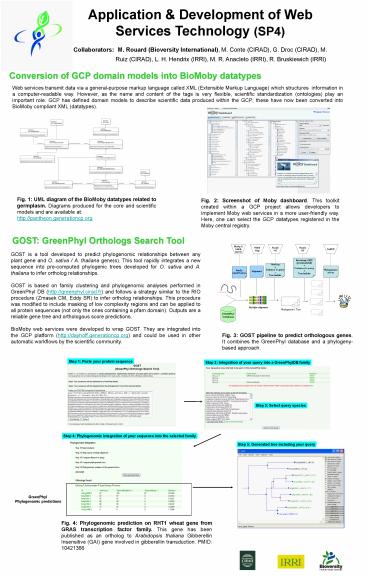
Fig. 2: Screenshot of Moby dashboard. ... Moby web services in a more user-friendly way. Here, one can select the GCP datatypes registered in the Moby central ... – PowerPoint PPT presentation
Number of Views:65
Avg rating:3.0/5.0
Title: Application
1
Application Development of Web Services
Technology (SP4)Collaborators M. Rouard
(Bioversity International), M. Conte (CIRAD), G.
Droc (CIRAD), M.
Ruiz (CIRAD), L. H. Hendrix (IRRI), M. R.
Anacleto (IRRI), R. Bruskiewich (IRRI)
Conversion of GCP domain models into BioMoby
datatypes
Web services transmit data via a general-purpose
markup language called XML (Extensible Markup
Language) which structures information in a
computer-readable way. However, as the name and
content of the tags is very flexible, scientific
standardization (ontologies) play an important
role. GCP has defined domain models to describe
scientific data produced within the GCP these
have now been converted into BioMoby compliant
XML (datatypes).
Fig. 1 UML diagram of the BioMoby datatypes
related to germplasm. Diagrams produced for the
core and scientific models and are available
at http//pantheon.generationcp.org
Fig. 2 Screenshot of Moby dashboard. This
toolkit created within a GCP project allows
developers to implement Moby web services in a
more user-friendly way. Here, one can select the
GCP datatypes registered in the Moby central
registry.
GOST GreenPhyl Orthologs Search Tool
GOST is a tool developed to predict phylogenomic
relationships between any plant gene and O.
sativa / A. thaliana gene(s). This tool rapidly
integrates a new sequence into pre-computed
phylogenic trees developed for O. sativa and A.
thaliana to infer ortholog relationships. GOST
is based on family clustering and phylogenomic
analyses performed in GreenPhyl DB
(http//greenphyl.cirad.fr) and follows a
strategy similar to the RIO procedure (Zmasek CM,
Eddy SR) to infer ortholog relationships. This
procedure was modified to include masking of low
complexity regions and can be applied to all
protein sequences (not only the ones containing a
pfam domain). Outputs are a reliable gene tree
and orthologous score predictions. BioMoby web
services were developed to wrap GOST. They are
integrated into the GCP platform
(http//dayhoff.generationcp.org) and could be
used in other automatic workflows by the
scientific community.
Fig. 3 GOST pipeline to predict orthologous
genes. It combines the GreenPhyl database and a
phylogeny-based approach.
Step 1 Paste your protein sequence
Step 2 Integration of your query into a
GreenPhylDB family
Step 3 Select query species
Step 4 Phylogenomic integration of your sequence
into the selected family
Step 5 Generated tree including your query
GreenPhyl Phylogenomic predictions
Fig. 4 Phylogenomic prediction on RHT1 wheat
gene from GRAS transcription factor family. This
gene has been published as an ortholog to
Arabidopsis thaliana Gibberellin Insensitive
(GAI) gene involved in gibberellin transduction.
PMID 10421366